Training and Evaluating Static Heatmap Regression Model for Multi-Instance and Single-Class Landmark Detetection (Plant Centroids)#
TODO
We will go through the following steps:
Setup environment#
!python -c "import landmarker" || pip install landmarker
Setup imports and variables#
import numpy as np
import torch
from torch.utils.data import DataLoader
from monai.transforms import (Compose, RandAffined, RandGaussianNoised, RandStdShiftIntensityd,
RandScaleIntensityd, RandAdjustContrastd, RandHistogramShiftd)
from tqdm import tqdm
device = 'cuda' if torch.cuda.is_available() else 'cpu'
Loading the dataset#
fn_keys = ('image',)
spatial_transformd = [RandAffined(fn_keys, prob=1,
rotate_range=(-np.pi/12, np.pi/12),
translate_range=(-10, 10),
scale_range=(-0.1, 0.1),
shear_range=(-0.1, 0.1)
)]
composed_transformd = Compose(spatial_transformd)
from landmarker.heatmap.generator import GaussianHeatmapGenerator
from landmarker.datasets import get_plant_centroids_landmark_datasets
ds_train, ds_test_A, ds_test_B, ds_test_C = get_plant_centroids_landmark_datasets("data", transform = composed_transformd, grayscale=False, store_imgs=False,
dim_img=(512,512))
100%|██████████| 1398/1398 [00:00<00:00, 5752.41it/s]
100%|██████████| 100/100 [00:00<00:00, 5978.11it/s]
100%|██████████| 275/275 [00:00<00:00, 6023.15it/s]
100%|██████████| 234/234 [00:00<00:00, 5655.99it/s]
Reading and extracting masks from 1398 images...
100%|██████████| 1398/1398 [00:04<00:00, 326.38it/s]
Reading and extracting masks from 100 images...
100%|██████████| 100/100 [00:00<00:00, 349.23it/s]
Reading and extracting masks from 275 images...
100%|██████████| 275/275 [00:00<00:00, 342.59it/s]
Reading and extracting masks from 234 images...
100%|██████████| 234/234 [00:00<00:00, 335.88it/s]
Constructing a heatmap generator#
from landmarker.heatmap.generator import GaussianHeatmapGenerator
heatmap_generator = GaussianHeatmapGenerator(
nb_landmarks=1,
sigmas=3,
gamma=None,
heatmap_size=(512, 512),
learnable=False, # If True, the heatmap generator will be trainable
device=device
)
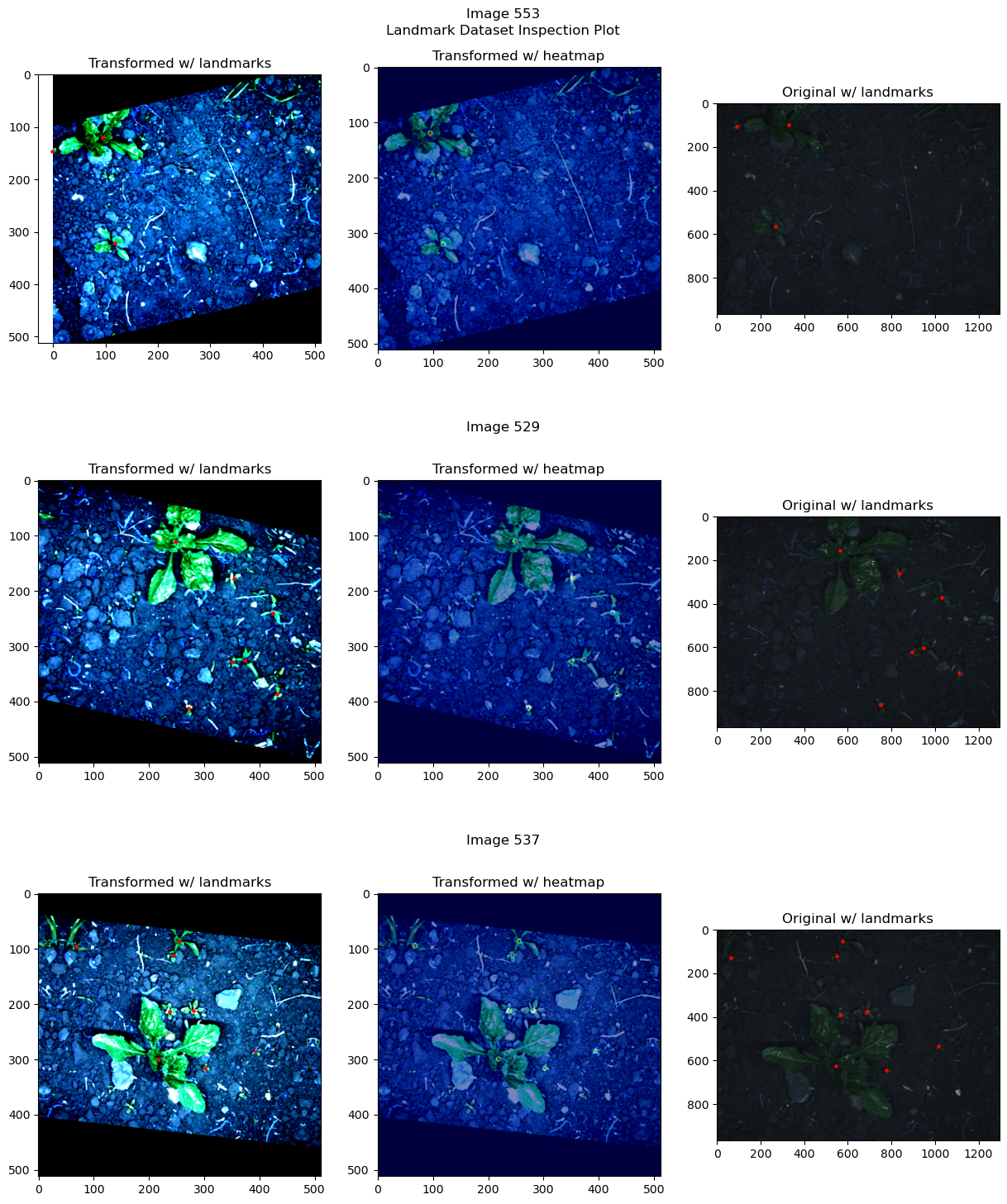
Inspecting the dataset#
from landmarker.visualize import inspection_plot
# Plot the first 3 images from the training set
heatmap_generator.device = "cpu" # because dataset tensors are still on cpu
inspection_plot(ds_train, np.random.randint(0, len(ds_train), 3),
heatmap_generator)
heatmap_generator.device = device # set the desired device back
/opt/conda/lib/python3.11/site-packages/torchvision/transforms/functional.py:1603: UserWarning: The default value of the antialias parameter of all the resizing transforms (Resize(), RandomResizedCrop(), etc.) will change from None to True in v0.17, in order to be consistent across the PIL and Tensor backends. To suppress this warning, directly pass antialias=True (recommended, future default), antialias=None (current default, which means False for Tensors and True for PIL), or antialias=False (only works on Tensors - PIL will still use antialiasing). This also applies if you are using the inference transforms from the models weights: update the call to weights.transforms(antialias=True).
warnings.warn(
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
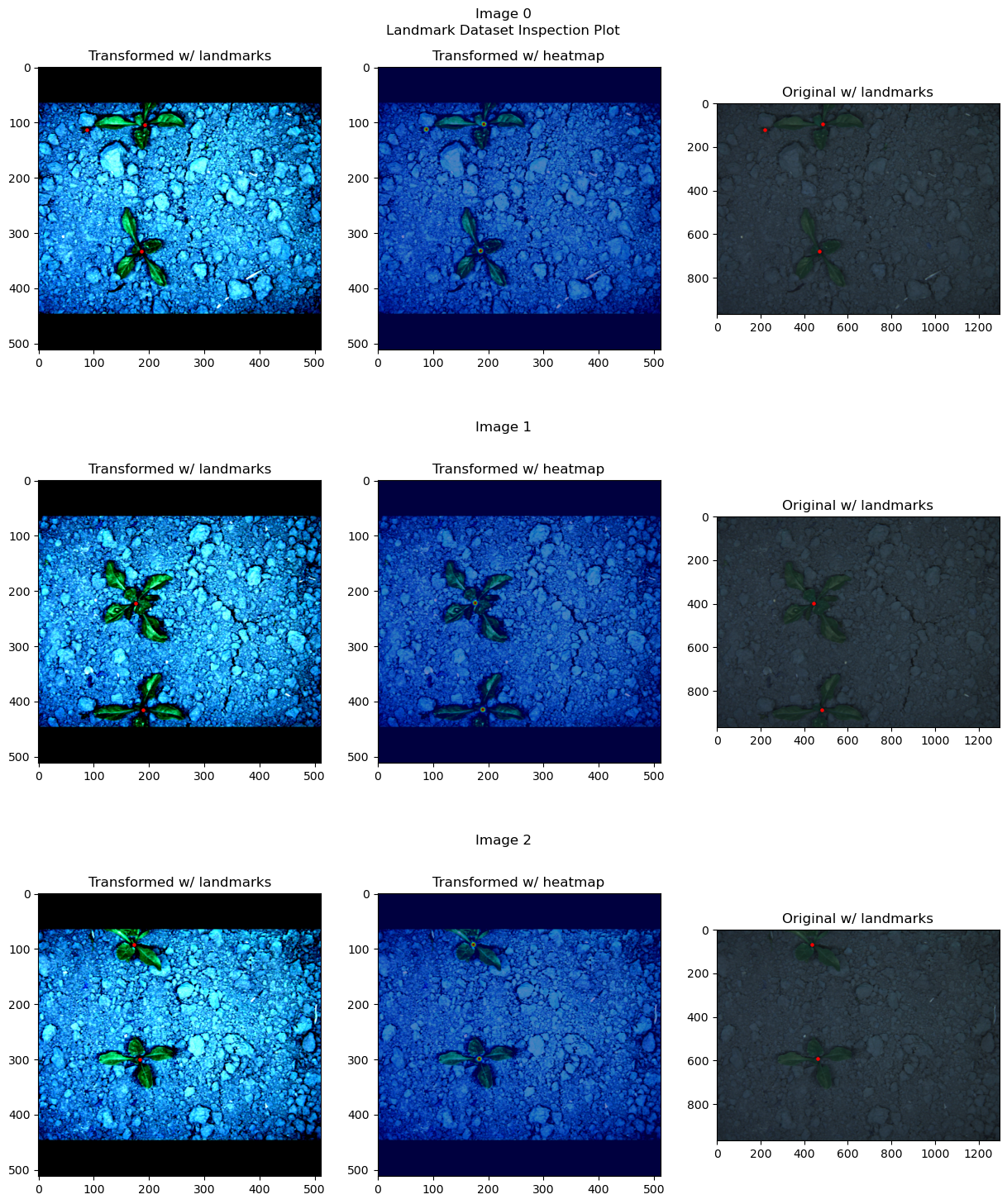
# Plot the first 3 images from the test1 set
heatmap_generator.device = "cpu" # because dataset tensors are still on cpu
inspection_plot(ds_test_A, range(3), heatmap_generator)
heatmap_generator.device = device # set the desired device back
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Training and initializing the SpatialConfiguration model#
Initializing the model, optimizer and loss function#
from torch import nn
from landmarker.models import get_model
from landmarker.train import EarlyStopping, SaveBestModel
model = get_model("UNet", in_channels=4, out_channels=1).to(device)
lr = 1e-4
batch_size = 4
epochs = 1000
optimizer = torch.optim.AdamW(model.parameters(), lr=lr, weight_decay=1e-3)
criterion = nn.BCEWithLogitsLoss()
early_stopping = EarlyStopping(patience=10, verbose=True)
save_best_model = SaveBestModel(verbose=True)
lr_scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, mode='min', factor=0.5,
patience=20, verbose=True, cooldown=10)
Setting the data loaders and split training set#
split_lengths = [0.8, 0.2]
ds_train_train, ds_train_val = torch.utils.data.random_split(ds_train, split_lengths)
train_loader = DataLoader(ds_train_train, batch_size=batch_size, shuffle=True, num_workers=0)
val_loader = DataLoader(ds_train_val, batch_size=batch_size, shuffle=False, num_workers=0)
test_A_loader = DataLoader(ds_test_A, batch_size=batch_size, shuffle=False, num_workers=0)
test_B_loader = DataLoader(ds_test_B, batch_size=batch_size, shuffle=False, num_workers=0)
test_C_loader = DataLoader(ds_test_C, batch_size=batch_size, shuffle=False, num_workers=0)
Training the model#
from landmarker.heatmap.decoder import heatmap_to_coord, heatmap_to_multiple_coord
from landmarker.metrics import point_error
from torch.nn.functional import sigmoid
from landmarker.metrics.metrics import multi_instance_point_error
def train_epoch(model, heatmap_generator, train_loader, criterion, optimizer, device):
running_loss = 0
model.train()
for i, (images, landmarks, _, _, _, _, _, _) in enumerate(tqdm(train_loader)):
images = images.to(device)
landmarks = landmarks.to(device)
optimizer.zero_grad()
outputs = model(images)
heatmaps = heatmap_generator(landmarks)
loss = criterion(outputs, heatmaps)
loss.backward()
optimizer.step()
running_loss += loss.item()
return running_loss / len(train_loader)
def val_epoch(model, heatmap_generator, val_loader, criterion, device):
eval_loss = 0
model.eval()
with torch.no_grad():
for _, (images, landmarks, _, _, _ , _, _,
_) in enumerate(tqdm(val_loader)):
images = images.to(device)
landmarks = landmarks.to(device)
outputs = model(images)
heatmaps = heatmap_generator(landmarks)
loss = criterion(outputs, heatmaps)
eval_loss += loss.item()
return eval_loss / len(val_loader)
def train(model, heatmap_generator, train_loader, val_loader, criterion, optimizer, device, epochs=1000):
for epoch in range(epochs):
ds_train.transform = composed_transformd
train_loss = train_epoch(model, heatmap_generator, train_loader, criterion, optimizer, device)
ds_train.transform = None
val_loss = val_epoch(model, heatmap_generator, val_loader, criterion, device)
print(f"Epoch {epoch+1}/{epochs} - Train loss: {train_loss:.4f} - Val loss: {val_loss:.4f}")
early_stopping(val_loss)
save_best_model(val_loss, model)
lr_scheduler.step(val_loss)
if early_stopping.early_stop:
print(f"Loading best model...")
model.load_state_dict(torch.load(save_best_model.path))
break
# train(model, heatmap_generator, train_loader, val_loader, criterion, optimizer, device,
# epochs=epochs)
model.load_state_dict(torch.load("model_static_unet_plant_centroids.pt"))
<All keys matched successfully>
Evaluating the model#
pred_landmarks = []
true_landmarks = []
dim_origs = []
pixel_spacings = []
paddings = []
tp = []
fp = []
fn = []
test_mpe = 0
test_tp = 0
test_fp = 0
test_fn = 0
threshold = 0.05
window = 9
model.eval()
model.to(device)
with torch.no_grad():
for i, (images, landmarks, affine_matrixs, _, _ ,
dim_orig, pixel_spacing, padding) in enumerate(tqdm(test_A_loader)):
images = images
landmarks = landmarks
outputs = model(images.to(device)).detach().cpu()
offset_coords = outputs.shape[1]-landmarks.shape[1]
pred_landmarks_list, _ = heatmap_to_multiple_coord(sigmoid(outputs), window=window,
threshold=threshold,
method="argmax")
pe_batch, tp_batch, fp_batch, fn_batch, pred_landmarks_torch = multi_instance_point_error(
true_landmarks=landmarks, pred_landmarks=pred_landmarks_list, dim=(512, 512),
dim_orig=dim_orig, pixel_spacing=pixel_spacing, padding=padding, reduction="none")
test_mpe += torch.nanmean(pe_batch).item()
test_tp += torch.nansum(tp_batch).item()
test_fp += torch.nansum(fp_batch).item()
test_fn += torch.nansum(fn_batch).item()
pred_landmarks.append(pred_landmarks_torch)
true_landmarks.append(landmarks)
dim_origs.append(dim_orig)
pixel_spacings.append(pixel_spacing)
paddings.append(padding)
tp.append(tp_batch)
fp.append(fp_batch)
fn.append(fn_batch)
test_mpe /= len(test_A_loader)
print(f"Test Mean PE: {test_mpe:.4f}")
print(f"Test TP: {test_tp:.4f}")
print(f"Test FP: {test_fp:.4f}")
print(f"Test FN: {test_fn:.4f}")
100%|██████████| 25/25 [05:55<00:00, 14.22s/it]
Test Mean PE: 13.3300
Test TP: 165.0000
Test FP: 12.0000
Test FN: 235.0000
from landmarker.metrics import sdr
sdr_test = sdr([4, 5, 10, 20], true_landmarks=torch.cat(true_landmarks, axis=0), pred_landmarks=torch.cat(pred_landmarks, axis=0),
dim=(512, 512), dim_orig=torch.cat(dim_origs, axis=0).int(), pixel_spacing=torch.cat(pixel_spacings, axis=0),
padding=torch.cat(paddings, axis=0))
for key in sdr_test:
print(f"SDR for {key}mm: {sdr_test[key]:.4f}")
SDR for 4mm: 6.0000
SDR for 5mm: 8.7500
SDR for 10mm: 21.7500
SDR for 20mm: 39.2500
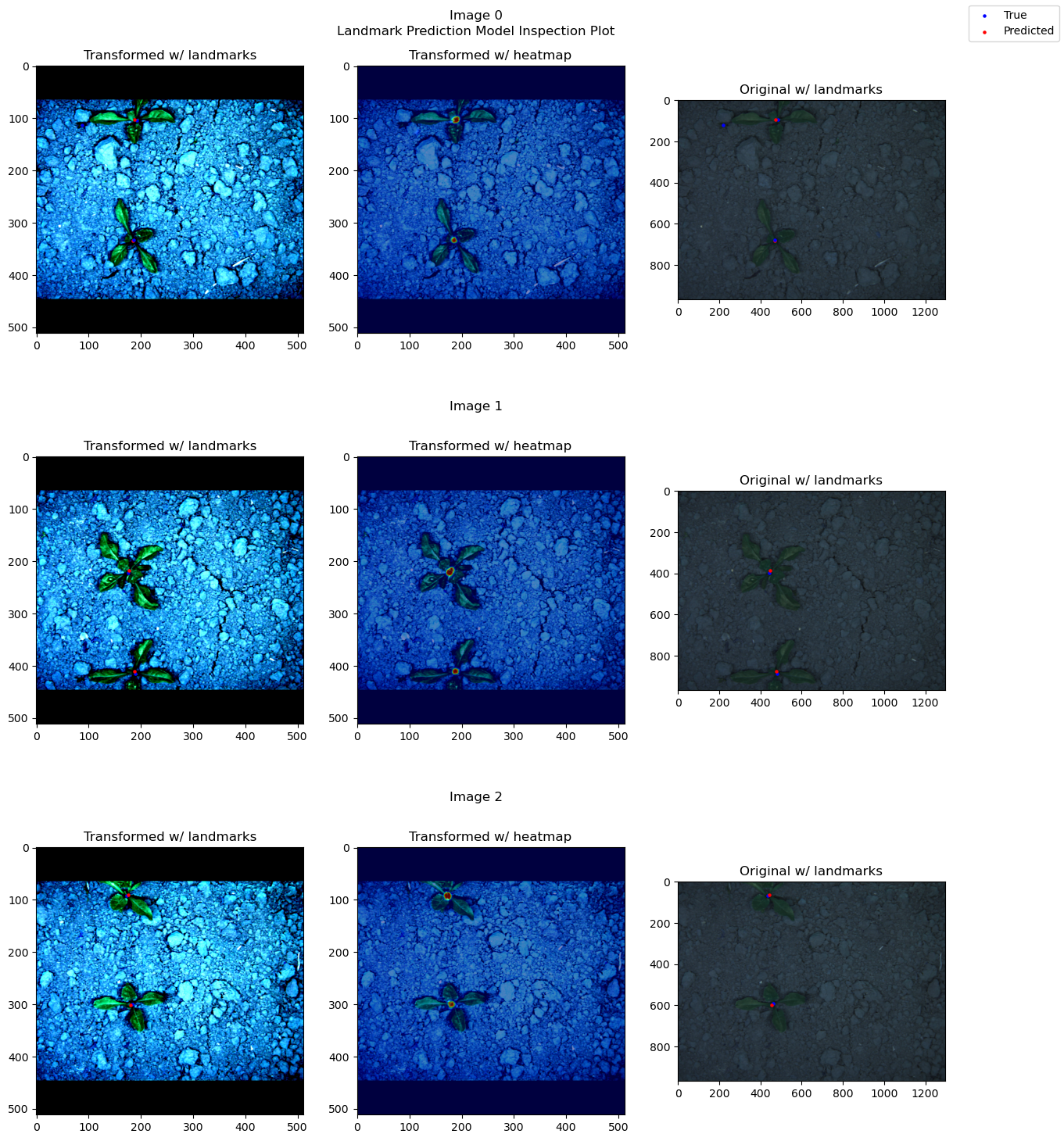
from landmarker.visualize.utils import prediction_inspect_plot_multi_instance
model.to("cpu")
prediction_inspect_plot_multi_instance(ds_test_A, model, range(3), threshold=threshold, activation=nn.Sigmoid())
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
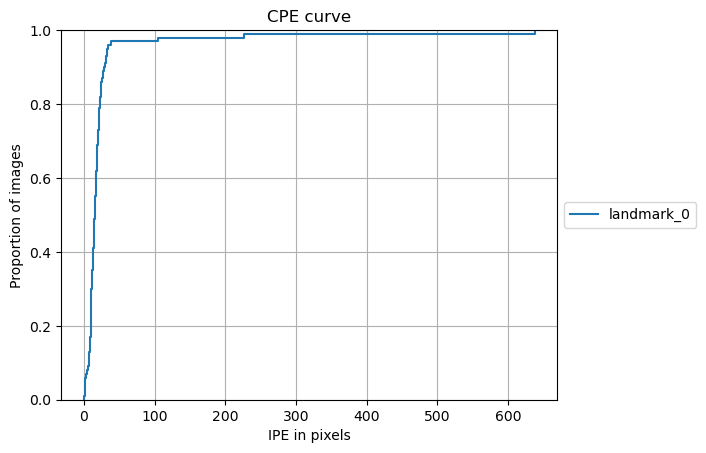
from landmarker.visualize import plot_cpe
plot_cpe(torch.cat(true_landmarks, axis=0), torch.cat(pred_landmarks, axis=0), dim=(512, 512),
dim_orig=torch.cat(dim_origs, axis=0).int(), pixel_spacing=torch.cat(pixel_spacings, axis=0),
padding=torch.cat(paddings, axis=0), class_names=ds_test_A.class_names,
group=False, title="CPE curve", save_path=None,
stat='proportion', unit='pixels', kind='ecdf')
from landmarker.visualize.evaluation import multi_instance_detection_report
multi_instance_detection_report(torch.cat(true_landmarks, axis=0), torch.cat(pred_landmarks, axis=0),
torch.cat(tp, axis=0), torch.cat(fp, axis=0), torch.cat(fn, axis=0), dim=(512, 512),
dim_orig=torch.cat(dim_origs, axis=0).int(), pixel_spacing=torch.cat(pixel_spacings, axis=0),
padding=torch.cat(paddings, axis=0), class_names=ds_test_A.class_names)
Detection report:
1# Instance detection statistics:
============================================================
Class TP FP FN TPR
------------------------------------------------------------
landmark_0 165.0 12.0 235.0 0.41
============================================================
2# Point-to-point error (PE) statistics:
======================================================================
Class Mean Median Std Min Max
----------------------------------------------------------------------
landmark_0 15.08 9.61 51.79 0.72 637.80
======================================================================
3# Success detection rate (SDR):
================================================================================
Class SDR (PE≤2mm) SDR (PE≤2.5mm) SDR (PE≤3mm) SDR (PE≤4mm)
--------------------------------------------------------------------------------
landmark_0 1.50 2.50 4.50 6.00
================================================================================
