Module tsflex.features
Feature extraction submodule.
Feature extraction guide
The following sections will explain the feature extraction module in detail.
Working example ✅
tsflex is built to be intuitive, so we encourage you to copy-paste this code and toy with some parameters!
This executable example creates a feature-collection that contains 2 features (skewness and minimum). Note that tsflex does not make any assumptions about the sampling rate of the time-series data.
import pandas as pd; import scipy.stats as ss; import numpy as np
from tsflex.features import FeatureDescriptor, FeatureCollection, FuncWrapper
# 1. -------- Get your time-indexed data --------
# Data contains 1 column; ["TMP"]
url = "https://github.com/predict-idlab/tsflex/raw/main/examples/data/empatica/"
data = pd.read_parquet(url + "tmp.parquet").set_index("timestamp")
# 2 -------- Construct your feature collection --------
fc = FeatureCollection(
feature_descriptors=[
FeatureDescriptor(
function=FuncWrapper(func=ss.skew, output_names="skew"),
series_name="TMP",
window="5min", stride="2.5min",
)
]
)
# -- 2.1. Add features to your feature collection
# NOTE: tsflex allows features to have different windows and strides
fc.add(FeatureDescriptor(np.min, "TMP", '2.5min', '2.5min'))
# 3 -------- Calculate features --------
fc.calculate(data=data, return_df=True) # which outputs:
| timestamp | TMP__amin__w=1m_s=30s | TMP__skew__w=2m_s=1m |
|---|---|---|
| 2017-06-13 14:23:13+02:00 | 27.37 | nan |
| 2017-06-13 14:23:43+02:00 | 27.37 | nan |
| 2017-06-13 14:24:13+02:00 | 27.43 | 10.8159 |
| 2017-06-13 14:24:43+02:00 | 27.81 | nan |
| 2017-06-13 14:25:13+02:00 | 28.23 | -0.0327893 |
| … | … | … |
Tip
More advanced feature-extraction examples can be found in these example notebooks
Getting started 🚀
The feature-extraction functionality of tsflex is provided by a FeatureCollection that contains FeatureDescriptors. The features are calculated (in a parallel manner) on the data that is passed to the feature collection.
Components
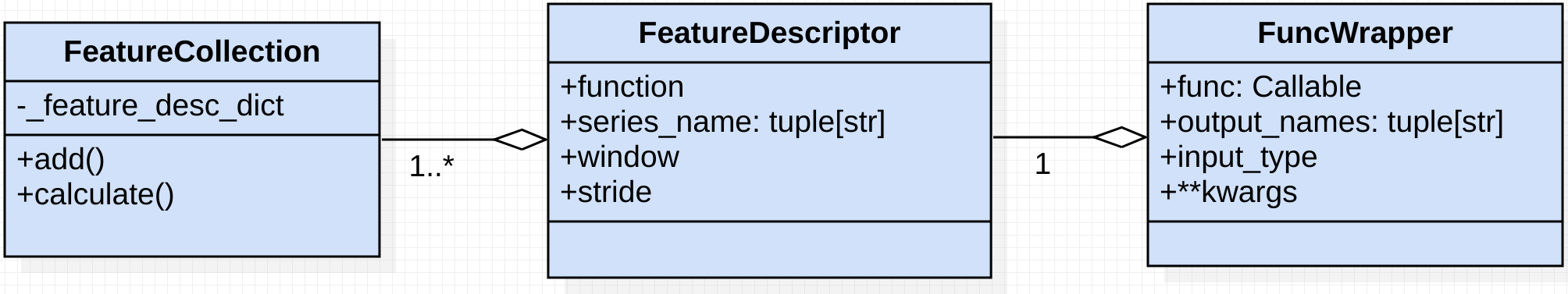
As shown above, there are 3 relevant classes for feature-extraction.
- FeatureCollection: serves as a registry, withholding the to-be-calculated features
- FeatureDescriptor: an instance of this class describes a feature.
Features are defined by:series_name: the names of the input series on which the feature-function will operatefunction: the Callable feature-function - e.g. np.meanwindow: the sample or time-based window - e.g. 200 or "2days"stride: the sample or time-based stride - e.g. 15 or "1hour"
- FuncWrapper: wraps Callable feature functions, and is intended for feature function configuration.
FuncWrappers are defined by:func: The wrapped feature-functionoutput_names: set custom and/or multiple feature output namesinput_type: define the feature its datatype; e.g., a pd.Series or np.array- **kwargs: additional keyword argument which are passed to
func
The snippet below shows how the FeatureCollection & FeatureDescriptor components work together:
import numpy as np; import scipy.stats as ss
from tsflex.features import FeatureDescriptor, FeatureCollection
# The FeatureCollection takes a List[FeatureDescriptor] as input
# There is no need for using a FuncWrapper when dealing with simple feature functions
fc = FeatureCollection(feature_descriptors=[
FeatureDescriptor(np.mean, "series_a", "1hour", "15min"),
FeatureDescriptor(ss.skew, "series_b", "3hours", "5min")
]
)
# We can still add features after instantiating.
fc.add(features=[FeatureDescriptor(np.std, "series_a", "1hour", "15min")])
# Calculate the features
fc.calculate(...)
Feature functions
A feature function needs to match this prototype:
function(*series: Union[np.ndarray, pd.Series], **kwargs)
-> Union[Any, List[Any]]
Hence, feature functions should take one (or multiple) arrays as first input. This can be followed by some keyword arguments.
The output of a feature function can be rather versatile (e.g., a float, integer, string, bool, … or a list thereof).
Note that the feature function may also take more than one series as input. In this case, the feature function should be wrapped in a FuncWrapper, with the input_type argument set to pd.Series.
In the advanced usage section, more info is given on these feature-function.
Multiple feature descriptors
Sometimes it can get overly verbose when the same feature is shared over multiple series, windows and/or strides. To solve this problem, we introduce the MultipleFeatureDescriptors. This component allows to create multiple feature descriptors for all the function - series_name(s) - window - stride combinations.
As shown in the example below, a MultipleFeatureDescriptors instance can be added a FeatureCollection.
import numpy as np; import scipy.stats as ss
from tsflex.features import FeatureDescriptor, FeatureCollection
from tsflex.features import MultipleFeatureDescriptors
# There is no need for using a FuncWrapper when dealing with simple feature functions
fc = FeatureCollection(feature_descriptors=[
FeatureDescriptor(np.mean, "series_a", "1hour", "15min"),
FeatureDescriptor(ss.skew, "series_b", "3hours", "5min"),
# Expands to a feature-descriptor list, withholding the combination of all
# The feature-window-stride arguments above.
MultipleFeatureDescriptors(
functions=[np.min, np.max, np.std, ss.skew],
series_names=["series_a", "series_b", "series_c"],
windows=["5min", "15min"],
strides=["1min","2min","3min"]
)
]
)
# Calculate the features
fc.calculate(...)
Output format
The output of the FeatureCollection its .calculate() is a (list of) sequence-indexed pd.DataFrames with column names:
<SERIES-NAME>__<FEAT-NAME>__w=<WINDOW>__s=<STRIDE>.
The column-name for the first feature defined in the snippet above will thus be series_a__std__w=1h__s=15m.
When the windows and strides are defined in a sample based-manner (which is mandatory for non datetime-indexed data), a possible output column would be series_a__std__w=100_s=15, where the window and stride are defined in samples and thus not in time-strings.
Note
You can find more information about the input data-formats in this section and read more about the (obvious) limitations in the next section.
Limitations ⚠️
It is important to note that there a still some, albeit logical, limitations regarding the supported data format.
These limitations are:
- Each
tsmust be in the flat/wide data format and they all need to have the same sequence index dtype, which needs to be sortable.- It just doesn't make sense to have a mix of different sequence index dtypes.
Imagine a FeatureCollection to which atswith apd.DatetimeIndexis passed, but atswith apd.RangeIndexis also passed. Both indexes aren't comparable, which thus is counterintuitive.
- It just doesn't make sense to have a mix of different sequence index dtypes.
- tsflex has no support for multi-indexes & multi-columns
- tsflex assumes that each
tshas a unique name. Hence no duplicatetsnames are allowed- Countermeasure: rename your
ts
- Countermeasure: rename your
Important notes 📢
We support various data-types. e.g. (np.float32, string-data, time-based data). However, it is the end-users responsibility to use a function which interplays nicely with the data its format.
Advanced usage 👀
Also take a look at the .reduce() and .serialize() methods.
Versatile functions
As explained above tsflex is rather versatile in terms of function input and output.
tsflex does not just allow one-to-one processing functions, but also many-to-one, one-to-many, and many-to-many functions are supported in a convenient way:
-
many-to-one; the feature function should- take multiple series as input
- output a single value
The function should (usually) not be wrapped in a
FuncWrapper.
Note that now theseries_nameargument requires a tuple of the ordered input series names.Example:
def abs_sum_diff(s1: np.array, s2: np.array) -> float:
min_len = min(len(s1), len(s2))
return np.sum(np.abs(s1[:min_len] - s2[:min_len]))
fd = FeatureDescriptor(
abs_sum_diff, series_name=("series_1", "series_2"),
window="5m", stride="2m30s",
)
-
one-to-many; the feature function should- take a single series as input
- output multiple values
The function should be wrapped in a
FuncWrapperto log its multiple output names.Example:
def abs_stats(s: np.array) -> Tuple[float]:
s_abs = np.abs(s)
return np.min(s_abs), np.max(s_abs), np.mean(s_abs), np.std(s_abs)
output_names = ["abs_min", "abs_max", "abs_mean", "abs_std"]
fd = FeatureDescriptor(
FuncWrapper(abs_stats, output_names=output_names),
series_name="series_1", window="5m", stride="2m30s",
)
-
many-to-many; the feature function should- take multiple series as input
- output multiple values
The function should be wrapped in a
FuncWrapperto log its multiple output names.Example:
def abs_stats_diff(s1: np.array, s2: np.array) -> Tuple[float]:
min_len = min(len(s1), len(s2))
s_abs_diff = np.sum(np.abs(s1[:min_len] - s2[:min_len]))
return np.min(s_abs_diff), np.max(s_abs_diff), np.mean(s_abs_diff)
output_names = ["abs_diff_min", "abs_diff_max", "abs_diff_mean"]
fd = FeatureDescriptor(
FuncWrapper(abs_stats_diff, output_names=output_names),
series_name=("series_1", "series_2"), window="5m", stride="2m30s",
)
Note
As visible in the feature function prototype, both np.array and pd.Series are supported function input types.
If your feature function requires pd.Series as input (instead of the default np.array), the function should be wrapped in a FuncWrapper with the input_type argument set to pd.Series.
An example of a function that leverages the pd.Series datatype:
def linear_trend_timewise(s: pd.Series):
# Get differences between each timestamp and the first timestamp in hour float
# Then convert to hours and reshape for linear regression
times_hours = np.asarray((s.index - s.index[0]).total_seconds() / 3600)
linReg = linregress(times_hours, s.values)
return linReg.slope, linReg.intercept, linReg.rvalue
fd = FeatureDescriptor(
FuncWrapper(
linear_trend_timewise,
["twise_regr_slope", "twise_regr_intercept", "twise_regr_r_value"],
input_type=pd.Series,
),
)
Multivariate-data
There are no assumptions made about the data its sequence-ranges. However, the end-user must take some things in consideration.
- By using the
bound_methodargument of.calculate(), the end-user can specify whether the "inner" or "outer" data-bounds will be used for generating the slice-ranges. - All
tsmust-have the same data-index dtype. this makes them comparable and allows for generating same-range slices on multivariate data.
Irregularly sampled data
Strided-rolling feature extraction on irregularly sampled data results in varying feature-segment sizes.
When using multivariate data, with either different sample rates or with an irregular data-rate, you cannot make the assumption that all windows will have the same length. Your feature extraction method should thus be:
- robust for varying length windows
- robust for (possible) empty windows
Tip
For conveniently creating such robust features we suggest using the make_robust function.
Note
A warning will be raised when irregular sampled data is observed.
In order to avoid this warning, the user should explicitly approve that there may be sparsity in the data by setting the approve_sparsity flag to True in the .calculate() method.
Logging
When a logging_file_path is passed to the FeatureCollection its .calculate() method, the execution times of the feature functions will be logged.
This is especially useful to identify which feature functions take a long time to compute.
Expand source code
"""Feature extraction submodule.
.. include:: ../../docs/pdoc_include/features.md
"""
__author__ = "Jonas Van Der Donckt, Jeroen Van Der Donckt, Emiel Deprost"
from .. import __pdoc__
from .feature import FeatureDescriptor, MultipleFeatureDescriptors
from .feature_collection import FeatureCollection
from .function_wrapper import FuncWrapper
from .logger import get_feature_logs, get_function_stats, get_series_names_stats
from .segmenter import StridedRollingFactory
__pdoc__["FuncWrapper.__call__"] = True
__all__ = [
"FeatureDescriptor",
"MultipleFeatureDescriptors",
"FeatureCollection",
"FuncWrapper",
"StridedRollingFactory",
"get_feature_logs",
"get_function_stats",
"get_series_names_stats",
]API reference of tsflex.features
.feature-
FeatureDescriptor and MultipleFeatureDescriptors class for creating time-series features.
.feature_collection-
FeatureCollection class for bookkeeping and calculation of time-series features …
.function_wrapper-
FuncWrapper class for object-oriented representation of a function.
.integrations-
Wrappers for seamless integration of feature functions from other packages.
.logger-
Contains the used variables and functions to provide logging functionality …
.segmenter-
Series segmentation submodule.
.utils-
Utility functions for more convenient feature extraction.
Functions
def get_feature_logs(logging_file_path)-
Expand source code
def get_feature_logs(logging_file_path: str) -> pd.DataFrame: """Get execution (time) info for each feature of a `FeatureCollection`. Parameters ---------- logging_file_path: str The file path where the logged messages are stored. This is the file path that is passed to the `FeatureCollection` its `calculate` method. Returns ------- pd.DataFrame A DataFrame with the features its function, input series names and (%) calculation duration. """ df = _parse_logging_execution_to_df(logging_file_path) df["duration"] = pd.to_timedelta(df["duration"], unit="s") return dfGet execution (time) info for each feature of a
FeatureCollection.Parameters
logging_file_path:str- The file path where the logged messages are stored. This is the file path that
is passed to the
FeatureCollectionitscalculatemethod.
Returns
pd.DataFrame- A DataFrame with the features its function, input series names and (%) calculation duration.
def get_function_stats(logging_file_path)-
Expand source code
def get_function_stats(logging_file_path: str) -> pd.DataFrame: """Get execution (time) statistics for each function. Parameters ---------- logging_file_path: str The file path where the logged messages are stored. This is the file path that is passed to the `FeatureCollection` its `calculate` method. Returns ------- pd.DataFrame A DataFrame with for each function (i.e., `function-(window,stride)`) combination the mean (time), std (time), sum (time), sum (% time), mean (% time),and number of executions. """ df = _parse_logging_execution_to_df(logging_file_path) # Get the sorted functions in a list to use as key for sorting the groups sorted_funcs = ( df.groupby(["function"]) .agg({"duration": ["mean"]}) .sort_values(by=("duration", "mean"), ascending=True) .index.to_list() ) def key_func(idx_level): # type: ignore[no-untyped-def] if all(idx in sorted_funcs for idx in idx_level): return [sorted_funcs.index(idx) for idx in idx_level] return idx_level return ( df.groupby(["function", "window", "stride"]) .agg( { "duration": ["sum", "mean", "std", "count"], "duration %": ["sum", "mean"], } ) .sort_index(key=key_func, ascending=False) )Get execution (time) statistics for each function.
Parameters
logging_file_path:str- The file path where the logged messages are stored. This is the file path that
is passed to the
FeatureCollectionitscalculatemethod.
Returns
pd.DataFrame- A DataFrame with for each function (i.e.,
function-(window,stride)) combination the mean (time), std (time), sum (time), sum (% time), mean (% time),and number of executions.
def get_series_names_stats(logging_file_path)-
Expand source code
def get_series_names_stats(logging_file_path: str) -> pd.DataFrame: """Get execution (time) statistics for each `key-(window,stride)` combination. Parameters ---------- logging_file_path: str The file path where the logged messages are stored. This is the file path that is passed to the `FeatureCollection` its `calculate` method. Returns ------- pd.DataFrame A DataFrame with for each function the mean (time), std (time), sum (time), sum (% time), mean (% time), and number of executions. """ df = _parse_logging_execution_to_df(logging_file_path) return ( df.groupby(["series_names", "window", "stride"]) .agg( { "duration": ["sum", "mean", "std", "count"], "duration %": ["sum", "mean"], } ) .sort_values(by=("duration", "sum"), ascending=False) )Get execution (time) statistics for each
key-(window,stride)combination.Parameters
logging_file_path:str- The file path where the logged messages are stored. This is the file path that
is passed to the
FeatureCollectionitscalculatemethod.
Returns
pd.DataFrame- A DataFrame with for each function the mean (time), std (time), sum (time), sum (% time), mean (% time), and number of executions.
Classes
class FeatureDescriptor (function, series_name, window=None, stride=None)-
Expand source code
class FeatureDescriptor(FrozenClass): """A FeatureDescriptor object, containing all feature information. Parameters ---------- function : Union[FuncWrapper, Callable] The function that calculates the feature(s). The prototype of the function should match: \n function(*series: Union[np.array, pd.Series]) -> Union[Any, List[Any]] Note that when the input type is ``pd.Series``, the function should be wrapped in a `FuncWrapper` with `input_type` = ``pd.Series``. series_name : Union[str, Tuple[str, ...]] The names of the series on which the feature function should be applied. This argument should match the `function` its input; \n * If `series_name` is a string (or tuple of a single string), then `function` should require just one series as input. * If `series_name` is a tuple of strings, then `function` should require `len(tuple)` series as input **and in exactly the same order** window : Union[float, str, pd.Timedelta], optional The window size. By default None. This argument supports multiple types: \n * If None, the `segment_start_idxs` and `segment_end_idxs` will need to be passed. * If the type is an `float` or an `int`, its value represents the series - its window **range** when a **non time-indexed** series is passed. - its window in **number of samples**, when a **time-indexed** series is passed (must then be and `int`) * If the window's type is a `pd.Timedelta`, the window size represents the window-time-range. The passed data **must have a time-index**. * If a `str`, it must represents a window-time-range-string. The **passed data must have a time-index**. .. Note:: - When the `segment_start_idxs` and `segment_end_idxs` are both passed to the `FeatureCollection.calculate` method, this window argument is ignored. Note that this is the only case when it is allowed to pass None for the window argument. - When the window argument is None, than the stride argument should be None as well (as it makes no sense to pass a stride value when the window is None). stride : Union[float, str, pd.Timedelta, List[Union[float, str, pd.Timedelta]]], optional The stride size(s). By default None. This argument supports multiple types: \n * If None, the stride will need to be passed to `FeatureCollection.calculate`. * If the type is an `float` or an `int`, its value represents the series - its stride **range** when a **non time-indexed** series is passed. - the stride in **number of samples**, when a **time-indexed** series is passed (must then be and `int`) * If the stride's type is a `pd.Timedelta`, the stride size represents the stride-time delta. The passed data **must have a time-index**. * If a `str`, it must represent a stride-time-delta-string. The **passed data must have a time-index**. \n * If a `List[Union[float, str, pd.Timedelta]]`, then the set intersection,of the strides will be used (e.g., stride=[2,3] -> index: 0, 2, 3, 4, 6, 8, 9, ...) .. Note:: - The stride argument of `FeatureCollection.calculate` takes precedence over this value when set (i.e., not None value for `stride` passed to the `calculate` method). - The stride argument should be None when the window argument is None (as it makes no sense to pass a stride value when the window is None). .. Note:: As described above, the `window-stride` argument can be sample-based (when using time-index series and int based arguments), but we do **not encourage** using this for `time-indexed` sequences. As we make the implicit assumption that the time-based data is sampled at a fixed frequency So only, if you're 100% sure that this is correct, you can safely use such arguments. Notes ----- * The `window` and `stride` argument should be either **both** numeric or ``pd.Timedelta`` (depending on de index datatype) - when `stride` is not None. * For each `function` - `input`(-series) - `window` - stride combination, one needs to create a distinct `FeatureDescriptor`. Hence it is more convenient to create a `MultipleFeatureDescriptors` when `function` - `window` - `stride` **combinations** should be applied on various input-series (combinations). * When `function` takes **multiple series** (i.e., arguments) as **input**, these are joined (based on the index) before applying the function. If the indexes of these series are not exactly the same, it might occur that not all series have exactly the same length! Hence, make sure that the `function` can deal with this! * For more information about the str-based time args, look into: [pandas time delta](https://pandas.pydata.org/pandas-docs/stable/user_guide/timedeltas.html#parsing){:target="_blank"} Raises ------ TypeError * Raised when the `function` is not an instance of Callable or FuncWrapper. * Raised when `window` and `stride` are not of exactly the same type (when `stride` is not None). See Also -------- StridedRolling: As the window-stride sequence conversion takes place there. """ def __init__( self, function: Union[FuncWrapper, Callable], series_name: Union[str, Tuple[str, ...]], window: Optional[Union[float, str, pd.Timedelta]] = None, stride: Optional[ Union[float, str, pd.Timedelta, List[Union[float, str, pd.Timedelta]]] ] = None, ): strides = sorted(set(to_list(stride))) # omit duplicate stride values if window is None: assert strides == [None], "stride must be None if window is None" self.series_name: Tuple[str, ...] = to_tuple(series_name) self.window = parse_time_arg(window) if isinstance(window, str) else window if strides == [None]: self.stride = None else: self.stride = [ parse_time_arg(s) if isinstance(s, str) else s for s in strides ] # Verify whether window and stride are either both sequence or time based dtype_set = set( AttributeParser.determine_type(v) for v in [self.window] + to_list(self.stride) ).difference([DataType.UNDEFINED]) if len(dtype_set) > 1: raise TypeError( f"a combination of window ({self.window} type={type(self.window)}) and" f" stride ({self.stride}) is not supported!" ) # Order of if statements is important (as FuncWrapper also is a Callable)! if isinstance(function, FuncWrapper): self.function: FuncWrapper = function elif isinstance(function, Callable): # type: ignore[arg-type] self.function: FuncWrapper = FuncWrapper(function) # type: ignore[no-redef] else: raise TypeError( "Expected feature function to be `Callable` or `FuncWrapper` but is a" f" {type(function)}." ) # Construct a function-string f_name = str(self.function) self._func_str: str = f"{self.__class__.__name__} - func: {f_name}" self._freeze() def get_required_series(self) -> List[str]: """Return all required series names for this feature descriptor. Return the list of series names that are required in order to execute the feature function. Returns ------- List[str] List of all the required series names. """ return list(set(self.series_name)) def get_nb_output_features(self) -> int: """Return the number of output features of this feature descriptor. Returns ------- int Number of output features. """ return len(self.function.output_names) def __repr__(self) -> str: """Representation string of Feature.""" return ( f"{self.__class__.__name__}({self.series_name}, {self.window}, " f"{self.stride})" )A FeatureDescriptor object, containing all feature information.
Parameters
function:Union[FuncWrapper, Callable]-
The function that calculates the feature(s). The prototype of the function should match:
function(*series: Union[np.array, pd.Series]) -> Union[Any, List[Any]]Note that when the input type is
pd.Series, the function should be wrapped in aFuncWrapperwithinput_type=pd.Series. series_name:Union[str, Tuple[str, …]]-
The names of the series on which the feature function should be applied. This argument should match the
functionits input;- If
series_nameis a string (or tuple of a single string), thenfunctionshould require just one series as input. - If
series_nameis a tuple of strings, thenfunctionshould requirelen(tuple)series as input and in exactly the same order
- If
window:Union[float, str, pd.Timedelta], optional-
The window size. By default None. This argument supports multiple types:
- If None, the
segment_start_idxsandsegment_end_idxswill need to be passed. - If the type is an
floator anint, its value represents the series- its window range when a non time-indexed series is passed.
- its window in number of samples, when a time-indexed series is
passed (must then be and
int)
- If the window's type is a
pd.Timedelta, the window size represents the window-time-range. The passed data must have a time-index. - If a
str, it must represents a window-time-range-string. The passed data must have a time-index.
Note
- When the
segment_start_idxsandsegment_end_idxsare both passed to the.calculate()method, this window argument is ignored. Note that this is the only case when it is allowed to pass None for the window argument. - When the window argument is None, than the stride argument should be None as well (as it makes no sense to pass a stride value when the window is None).
- If None, the
stride:Union[float, str, pd.Timedelta, List[Union[float, str, pd.Timedelta]]], optional-
The stride size(s). By default None. This argument supports multiple types:
- If None, the stride will need to be passed to
.calculate(). - If the type is an
floator anint, its value represents the series- its stride range when a non time-indexed series is passed.
- the stride in number of samples, when a time-indexed series
is passed (must then be and
int)
- If the stride's type is a
pd.Timedelta, the stride size represents the stride-time delta. The passed data must have a time-index. -
If a
str, it must represent a stride-time-delta-string. The passed data must have a time-index. -
If a
List[Union[float, str, pd.Timedelta]], then the set intersection,of the strides will be used (e.g., stride=[2,3] -> index: 0, 2, 3, 4, 6, 8, 9, …)
Note
- The stride argument of
.calculate()takes precedence over this value when set (i.e., not None value forstridepassed to thecalculatemethod). - The stride argument should be None when the window argument is None (as it makes no sense to pass a stride value when the window is None).
- If None, the stride will need to be passed to
Note
As described above, the
window-strideargument can be sample-based (when using time-index series and int based arguments), but we do not encourage using this fortime-indexedsequences. As we make the implicit assumption that the time-based data is sampled at a fixed frequency So only, if you're 100% sure that this is correct, you can safely use such arguments.Notes
- The
windowandstrideargument should be either both numeric orpd.Timedelta(depending on de index datatype) - whenstrideis not None. - For each
function-input(-series) -window- stride combination, one needs to create a distinctFeatureDescriptor. Hence it is more convenient to create aMultipleFeatureDescriptorswhenfunction-window-stridecombinations should be applied on various input-series (combinations). - When
functiontakes multiple series (i.e., arguments) as input, these are joined (based on the index) before applying the function. If the indexes of these series are not exactly the same, it might occur that not all series have exactly the same length! Hence, make sure that thefunctioncan deal with this! - For more information about the str-based time args, look into: pandas time delta
Raises
TypeError-
- Raised when the
functionis not an instance of Callable or FuncWrapper. - Raised when
windowandstrideare not of exactly the same type (whenstrideis not None).
- Raised when the
See Also
StridedRolling- As the window-stride sequence conversion takes place there.
Ancestors
- tsflex.utils.classes.FrozenClass
Methods
def get_required_series(self)-
Expand source code
def get_required_series(self) -> List[str]: """Return all required series names for this feature descriptor. Return the list of series names that are required in order to execute the feature function. Returns ------- List[str] List of all the required series names. """ return list(set(self.series_name))Return all required series names for this feature descriptor.
Return the list of series names that are required in order to execute the feature function.
Returns
List[str]- List of all the required series names.
def get_nb_output_features(self)-
Expand source code
def get_nb_output_features(self) -> int: """Return the number of output features of this feature descriptor. Returns ------- int Number of output features. """ return len(self.function.output_names)Return the number of output features of this feature descriptor.
Returns
int- Number of output features.
class MultipleFeatureDescriptors (functions, series_names, windows=None, strides=None)-
Expand source code
class MultipleFeatureDescriptors: """Create a MultipleFeatureDescriptors object. Create a list of features from **all** combinations of the given parameter lists. Total number of created `FeatureDescriptor`s will be: len(func_inputs)*len(functions)*len(windows)*len(strides). Parameters ---------- functions : Union[FuncWrapper, Callable, List[Union[FuncWrapper, Callable]]] The functions, can be either of both types (even in a single array). series_names : Union[str, Tuple[str, ...], List[str], List[Tuple[str, ...]]] The names of the series on which the feature function should be applied. * If `series_names` is a (list of) string (or tuple of a single string), then each `function` should require just one series as input. * If `series_names` is a (list of) tuple of strings, then each `function` should require `len(tuple)` series as input. A `list` implies that multiple multiple series (combinations) will be used to extract features from; \n * If `series_names` is a string or a tuple of strings, then `function` will be called only once for the series of this argument. * If `series_names` is a list of either strings or tuple of strings, then `function` will be called for each entry of this list. .. Note:: when passing a list as `series_names`, all items in this list should have the same type, i.e, either \n * all a str * or, all a tuple _with same length_.\n And perfectly match the func-input size. windows : Union[float, str, pd.Timedelta, List[Union[float, str, pd.Timedelta]]] All the window sizes. strides : Union[float, str, pd.Timedelta, None, List[Union[float, str, pd.Timedelta]]], optional All the strides. By default None. Note ---- The `windows` and `strides` argument should be either both numeric or ``pd.Timedelta`` (depending on de index datatype) - when `strides` is not None. """ def __init__( self, functions: Union[FuncWrapper, Callable, List[Union[FuncWrapper, Callable]]], series_names: Union[str, Tuple[str, ...], List[str], List[Tuple[str, ...]]], windows: Optional[ Union[float, str, pd.Timedelta, List[Union[float, str, pd.Timedelta]]] ] = None, strides: Optional[ Union[float, str, pd.Timedelta, List[Union[float, str, pd.Timedelta]]] ] = None, ): # Cast functions to FuncWrapper, this avoids creating multiple # FuncWrapper objects for the same function in the FeatureDescriptor def to_func_wrapper(f: Callable) -> FuncWrapper: return f if isinstance(f, FuncWrapper) else FuncWrapper(f) functions = [to_func_wrapper(f) for f in to_list(functions)] # Convert the series names to list of tuples series_names = [to_tuple(names) for names in to_list(series_names)] # Assert that function inputs (series) all have the same length assert all( len(series_names[0]) == len(series_name_tuple) for series_name_tuple in series_names ) # Convert the other types to list windows = to_list(windows) self.feature_descriptions: List[FeatureDescriptor] = [] # Iterate over all combinations combinations = [functions, series_names, windows] for function, series_name, window in itertools.product(*combinations): # type: ignore[call-overload] self.feature_descriptions.append( FeatureDescriptor(function, series_name, window, strides) )Create a MultipleFeatureDescriptors object.
Create a list of features from all combinations of the given parameter lists. Total number of created
FeatureDescriptors will be:len(func_inputs)*len(functions)*len(windows)*len(strides).Parameters
functions:Union[FuncWrapper, Callable, List[Union[FuncWrapper, Callable]]]- The functions, can be either of both types (even in a single array).
series_names:Union[str, Tuple[str, …], List[str], List[Tuple[str, …]]]-
The names of the series on which the feature function should be applied.
- If
series_namesis a (list of) string (or tuple of a single string), then eachfunctionshould require just one series as input. - If
series_namesis a (list of) tuple of strings, then eachfunctionshould requirelen(tuple)series as input.
A
listimplies that multiple multiple series (combinations) will be used to extract features from;- If
series_namesis a string or a tuple of strings, thenfunctionwill be called only once for the series of this argument. - If
series_namesis a list of either strings or tuple of strings, thenfunctionwill be called for each entry of this list.
Note
when passing a list as
series_names, all items in this list should have the same type, i.e, either- all a str
- or, all a tuple with same length.
And perfectly match the func-input size.
- If
windows:Union[float, str, pd.Timedelta, List[Union[float, str, pd.Timedelta]]]- All the window sizes.
strides:Union[float, str, pd.Timedelta, None, List[Union[float, str, pd.Timedelta]]], optional- All the strides. By default None.
Note
The
windowsandstridesargument should be either both numeric orpd.Timedelta(depending on de index datatype) - whenstridesis not None. class FeatureCollection (feature_descriptors=None)-
Expand source code
class FeatureCollection: """Create a FeatureCollection. Parameters ---------- feature_descriptors : Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection, List[Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection]]], optional Initial (list of) feature(s) to add to collection, by default None Notes ----- * The `series_name` property of the `FeatureDescriptor`s should **not withhold a "|" character**, since "|" is used to join the series names of features which use multiple series as input).<br> e.g.<br> * `ACC|x` is **not** allowed as series name, as this is ambiguous and could represent that this feature is constructed with a combination of the `ACC` and `x` signal.<br> Note that `max|feat` is allowed as feature output name. * Both the `series_name` and `output_name` property of the `FeatureDescriptor`s **should not withhold "__"** in its string representations. This constraint is mainly made for readability purposes. The two statements above will be asserted """ def __init__( self, feature_descriptors: Optional[ Union[ FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection, Sequence[ Union[ FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection ] ], ] ] = None, ): # The feature collection is a dict with keys of type: # tuple(tuple(str), float OR pd.timedelta) # The outer tuple's values correspond to (series_key(s), window) self._feature_desc_dict: Dict[ Tuple[Tuple[str, ...], Union[float, pd.Timedelta]], List[FeatureDescriptor] ] = {} if feature_descriptors: self.add(feature_descriptors) def get_required_series(self) -> List[str]: """Return all required series names for this feature collection. Return the list of series names that are required in order to calculate all the features (defined by the `FeatureDescriptor` objects) of this feature collection. Returns ------- List[str] List of all the required series names. """ return list( set(flatten([fr_key[0] for fr_key in self._feature_desc_dict.keys()])) ) def get_nb_output_features(self) -> int: """Return the number of output features in this feature collection. Returns ------- int The number of output features in this feature collection. """ fd_list: Iterable[FeatureDescriptor] = flatten(self._feature_desc_dict.values()) return sum(fd.get_nb_output_features() for fd in fd_list) def _get_nb_output_features_without_window(self) -> int: """Return the number of output features in this feature collection, without using the window as a unique identifier. This is relevant for when the window value(s) are overridden by passing `segment_start_idxs` and `segment_end_idxs` to the `calculate` method. Returns ------- int: The number of output features in this feature collection without using the window as a unique identifier. """ return len( set( (series, o) for (series, _), fd_list in self._feature_desc_dict.items() for fd in fd_list for o in fd.function.output_names ) ) def _get_nb_feat_funcs(self) -> int: return sum(map(len, self._feature_desc_dict.values())) @staticmethod def _get_collection_key( feature: FeatureDescriptor, ) -> Tuple[Tuple[str, ...], Union[pd.Timedelta, float, None]]: # Note: `window` property can be either a pd.Timedelta or a float or None # assert feature.window is not None return feature.series_name, feature.window def _check_feature_descriptors( self, skip_none: bool, calc_stride: Optional[Union[float, pd.Timedelta, None]] = None, ) -> None: """Verify whether all added FeatureDescriptors imply the same-input data type. If this condition is not met, a warning will be raised. Parameters ---------- skip_none: bool Whether to include None stride values in the checks. calc_stride: Union[float, pd.Timedelta, None], optional The `FeatureCollection.calculate` its stride argument, by default None. This stride takes precedence over a `FeatureDescriptor` its stride when it is not None. """ dtype_set = set() for series_names, win in self._feature_desc_dict.keys(): for fd in self._feature_desc_dict[(series_names, win)]: stride = calc_stride if calc_stride is not None else fd.stride if skip_none and stride is None: dtype_set.add(AttributeParser.determine_type(win)) else: dtype_set.add( AttributeParser.determine_type([win] + to_list(stride)) ) if len(dtype_set) > 1: warnings.warn( "There are multiple FeatureDescriptor window-stride " + f"datatypes present in this FeatureCollection, i.e.: {dtype_set}", category=RuntimeWarning, ) def _add_feature(self, feature: FeatureDescriptor) -> None: """Add a `FeatureDescriptor` instance to the collection. Parameters ---------- feature : FeatureDescriptor The feature that will be added to this feature collection. """ # Check whether the `|` is not present in the series assert not any("|" in s_name for s_name in feature.get_required_series()) # Check whether the '__" is not present in the series and function output names assert not any( "__" in output_name for output_name in feature.function.output_names ) assert not any("__" in s_name for s_name in feature.get_required_series()) series_win_stride_key = self._get_collection_key(feature) if series_win_stride_key in self._feature_desc_dict.keys(): added_output_names = flatten( f.function.output_names for f in self._feature_desc_dict[series_win_stride_key] ) # Check that not a feature with the same output_name(s) is already added # for the series_win_stride_key assert not any( output_name in added_output_names for output_name in feature.function.output_names ), f"Feature with output name(s) {feature.function.output_names} duplicated" self._feature_desc_dict[series_win_stride_key].append(feature) else: self._feature_desc_dict[series_win_stride_key] = [feature] def add( self, features: Union[ FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection, Sequence[ Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection] ], ], ) -> None: """Add feature(s) to the FeatureCollection. Parameters ---------- features : Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection, List[Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection]]] Feature(s) (containers) whose contained features will be added. Raises ------ TypeError Raised when an item within `features` is not an instance of [`MultipleFeatureDescriptors`, `FeatureDescriptors`, `FeatureCollection`]. """ # Convert to list if necessary features = to_list(features) for feature in features: if isinstance(feature, MultipleFeatureDescriptors): self.add(feature.feature_descriptions) elif isinstance(feature, FeatureDescriptor): self._add_feature(feature) elif isinstance(feature, FeatureCollection): # List needs to be flattened self.add(list(flatten(feature._feature_desc_dict.values()))) else: raise TypeError(f"type: {type(feature)} is not supported - {feature}") # After adding the features, check whether the descriptors are compatible self._check_feature_descriptors(skip_none=True) @staticmethod def _executor_stroll(idx: int) -> pd.DataFrame: """Executor function for the StridedRolling.apply_func method. Strided rolling feature calculation occurs when either; - a `window` and `stride` argument are stored in the `FeatureDescriptor` object - the `window` is stored in the `FeatureDescriptor` object and the `stride` argument is passed to the `calculate` method, potentially overriding the `stride` - segment indices are passed to the `calculate` method - a `group_by_consecutive` argument is passed to the `calculate` method (since we calculate the segment indices for the consecutive groups) This method uses the global `get_stroll_func` function, which returns the StridedRolling object and the function that needs to be applied to the StridedRolling object. Using a global function is necessary to facilitate multiprocessing. """ # Uses the global get_stroll_func stroll, function = get_stroll_func(idx) return stroll.apply_func(function) # execution time is logged in apply_func @staticmethod def _executor_grouped(idx: int) -> pd.DataFrame: """Executor function for grouped feature calculation. Grouped feature calculation occurs when either; - a `group_by_all` argument is passed to the `calculate` method - a `DataFrameGroupBy` is passed as `data` argument to the `calculate` method Note that passing a `group_by_consecutive` argument to the `calculate` method will not use this executor function, but will use the `_executor_stroll` as executor function (since we calculate the segment indices for the consecutive groups). This method uses the global `get_group_func` function, which returns a pd.DataFrame (containing only the necessary data for the function) and the function that needs to be applied to the pd.DataFrame. In addition, the global `group_indices` and `group_id_name` are used to access the grouped data and the group id name respectively. Using a global function is necessary to facilitate multiprocessing. """ # Uses the global get_group_func, group_indices, and group_id_name data, function = get_group_func(idx) group_ids = group_indices.keys() # group_ids are the keys of the group_indices cols_tuple = tuple(data.columns.values) t_start = time.perf_counter() # Wrap the function to handle multiple inputs and convert the inputs to numpy # array if necessary f = function if function.input_type is np.array: def f(x: pd.DataFrame) -> Any: # pass the inputs as positional arguments of numpy array type return function(*[x[c].values for c in cols_tuple]) else: # function.input_type is pd.Series def f(x: pd.DataFrame) -> Any: # pass the inputs as positional arguments of pd.Series type return function(*[x[c] for c in cols_tuple]) # Function execution over the grouped data (accessed by using the group_indices) out = np.array(list(map(f, [data.iloc[idx] for idx in group_indices.values()]))) # Aggregate function output in a dictionary output_names = [ StridedRolling.construct_output_index( cols_tuple, feat_name, win_str="manual" ) for feat_name in function.output_names ] feat_out = _process_func_output(out, group_ids, output_names, str(function)) # Log the function execution time _log_func_execution( t_start, function, cols_tuple, "manual", "manual", output_names ) return pd.DataFrame(feat_out, index=group_ids).rename_axis(index=group_id_name) def _stroll_feat_generator( self, series_dict: Dict[str, pd.Series], calc_stride: Union[List[Union[float, pd.Timedelta]], None], segment_start_idxs: Union[np.ndarray, None], segment_end_idxs: Union[np.ndarray, None], start_idx: Any, end_idx: Any, window_idx: str, include_final_window: bool, approve_sparsity: bool, ) -> Callable[[int], Tuple[StridedRolling, FuncWrapper]]: # --- Future work --- # We could also make the StridedRolling creation multithreaded # Very low priority because the STROLL __init__ is rather efficient! keys_wins_strides = list(self._feature_desc_dict.keys()) lengths = np.cumsum( [len(self._feature_desc_dict[k]) for k in keys_wins_strides] ) def get_stroll_function(idx: int) -> Tuple[StridedRolling, FuncWrapper]: key_idx = np.searchsorted(lengths, idx, "right") # right bc idx starts at 0 key, win = keys_wins_strides[key_idx] feature = self._feature_desc_dict[keys_wins_strides[key_idx]][ idx - lengths[key_idx] ] stride = feature.stride if calc_stride is None else calc_stride function: FuncWrapper = feature.function # The factory method will instantiate the right StridedRolling object stroll_arg_dict = dict( data=[series_dict[k] for k in key], window=win, strides=stride, segment_start_idxs=segment_start_idxs, segment_end_idxs=segment_end_idxs, start_idx=start_idx, end_idx=end_idx, window_idx=window_idx, include_final_window=include_final_window, approve_sparsity=approve_sparsity, func_data_type=function.input_type, ) stroll = StridedRollingFactory.get_segmenter(**stroll_arg_dict) return stroll, function return get_stroll_function def _group_feat_generator( self, grouped_df: pd.api.typing.DataFrameGroupBy, ) -> Callable[[int], Tuple[pd.api.typing.DataFrameGroupBy, FuncWrapper,],]: """Return a function that returns the necessary columns of the grouped data and the function that needs to be applied to the grouped data. Note that the function does not return groups, but rather the necessary columns of the grouped data (i.e. the data on which the function needs to be applied). To access the groups, the global `group_indices` and `group_id_name` are used. """ keys_wins = list(self._feature_desc_dict.keys()) lengths = np.cumsum([len(self._feature_desc_dict[k]) for k in keys_wins]) def get_group_function( idx: int, ) -> Tuple[pd.api.typing.DataFrameGroupBy, FuncWrapper,]: key_idx = np.searchsorted(lengths, idx, "right") # right bc idx starts at 0 key, win = keys_wins[key_idx] feature = self._feature_desc_dict[keys_wins[key_idx]][ idx - lengths[key_idx] ] function: FuncWrapper = feature.function return grouped_df.obj[list(key)], function return get_group_function def _check_no_multiple_windows(self, error_case: str) -> None: """Check whether there are no multiple windows in the feature collection. Parameters ---------- error_case : str The case in which no multiple windows are allowed. """ assert ( self._get_nb_output_features_without_window() == self.get_nb_output_features() ), ( error_case + "; each output name - series_input combination can only have 1 window" + " (or None)" ) @staticmethod def _data_to_series_dict( data: Union[pd.DataFrame, pd.Series, List[Union[pd.Series, pd.DataFrame]]], required_series: List[str], ) -> Dict[str, pd.Series]: series_dict: Dict[str, pd.Series] = {} for s in to_series_list(data): if not s.index.is_monotonic_increasing: warnings.warn( f"The index of series '{s.name}' is not monotonic increasing. " + "The series will be sorted by the index.", RuntimeWarning, ) s = s.sort_index(ascending=True, inplace=False, ignore_index=False) # Assert the assumptions we make! assert s.index.is_monotonic_increasing if s.name in required_series: series_dict[str(s.name)] = s return series_dict @staticmethod def _process_segment_idxs( segment_idxs: Union[list, np.ndarray, pd.Series, pd.Index] ) -> np.ndarray: if hasattr(segment_idxs, "values"): segment_idxs = segment_idxs.values segment_idxs = np.asarray(segment_idxs) if segment_idxs.ndim > 1: segment_idxs = segment_idxs.squeeze() # remove singleton dimensions return segment_idxs @staticmethod def _group_by_all( series_dict: Dict[str, pd.Series], col_name: str ) -> pd.api.typing.DataFrameGroupBy: """Group all `column_name` values and return the grouped data. GroupBy ignores all rows with NaN values for the column on which we group. Parameters ---------- series_dict : Dict[str, pd.Series] Input data. col_name : str The column name on which the grouping will need to take place. Returns ------- pd.api.typing.DataFrameGroupBy A `DataFrameGroupBy` object, with the group names as keys and the indices as values. """ df = pd.DataFrame(series_dict) assert col_name in df.columns # Check if there are nan values in the column on which we group if df[col_name].isna().any(): warnings.warn( f"NaN values were found in the column '{col_name}' (when expanding the " + f"data to a pd.DataFrame which contains {df.columns}. " + "Rows with NaN values for the grouping column will be ignored.", RuntimeWarning, ) # GroupBy ignores all rows with NaN values for the column on which we group return df.groupby(col_name) def _calculate_group_by_all( self, grouped_data: pd.api.typing.DataFrameGroupBy, return_df: bool, show_progress: bool, n_jobs: Optional[int], f_handler: Optional[logging.FileHandler], ) -> Union[List[pd.DataFrame], pd.DataFrame]: """Calculate features on each group of the grouped data. Parameters ---------- grouped_data : pd.api.typing.DataFrameGroupBy The grouped data. return_df: bool, optional Whether the output needs to be a DataFrame or a list thereof. show_progress: bool, optional Whether to show a progress bar. n_jobs: int, optional The number of jobs to run in parallel. f_handler: logging.FileHandler, optional The file handler that is used to log the function execution times. .. Note:: Is comparable to following pseudo-SQL code: ```sql SELECT func(x) FROM `data` GROUP BY ... ``` where `func` is the FeatureDescriptor function and `x` is the name on which the FeatureDescriptor operates. The group by is already done by passing a `DataFrameGroupBy` object to this method. """ global group_indices, group_id_name, get_group_func group_indices = grouped_data.indices # dict - group_id as key; indices as value # since in future versions of pandas grouper will be deprecated group_attr = "_grouper" if hasattr(grouped_data, "_grouper") else "grouper" group_id_name = getattr(grouped_data, group_attr).names # name of group col(s) get_group_func = self._group_feat_generator(grouped_data) # sort_output_index can be set to False, since we want to keep the same order as # the group_indices return self._calculate_feature_list( self._executor_grouped, n_jobs, show_progress, return_df, False, f_handler ) @staticmethod def _group_by_consecutive( df: Union[pd.Series, pd.DataFrame], col_name: Optional[str] = None ) -> pd.DataFrame: """Group consecutive `col_name` values in a single DataFrame. This is especially useful if you want to represent sparse data in a more compact format. Parameters ---------- df : Union[pd.Series, pd.DataFrame] Input data. col_name : str, optional If a dataFrame is passed, you will need to specify the `col_name` on which the consecutive-grouping will need to take place. Returns ------- pd.DataFrame A new `DataFrame` view, with columns: [`start`, `end`, `col_name`], representing the start- and endtime of the consecutive range, and the col_name's consecutive values. """ if type(df) == pd.Series: col_name = df.name df = df.to_frame() assert col_name in df.columns assert col_name not in [ "start", "end", ], "Grouping column cannot be 'start' or 'end'" # Check if there are nan values in the column on which we group if df[col_name].isna().any(): warnings.warn( f"NaN values were found in the column '{col_name}' (when expanding the " + f"data to a pd.DataFrame which contains {df.columns}. " + "Rows with NaN values for the grouping column will be ignored.", RuntimeWarning, ) # Drop all rows with NaN values for the column on which we group df.dropna(subset=[col_name], inplace=True) df_cum = ( (df[col_name] != df[col_name].shift(1)) .astype("int") .cumsum() .rename("value_grp") .to_frame() ) df_cum["sequence_idx"] = df.index df_cum[col_name] = df[col_name] df_cum_grouped = df_cum.groupby("value_grp") df_grouped = pd.DataFrame( { "start": df_cum_grouped["sequence_idx"].first(), "end": df_cum_grouped["sequence_idx"].last(), col_name: df_cum_grouped[col_name].first(), } ).reset_index(drop=True) return df_grouped def _calculate_group_by_consecutive( # type: ignore[no-untyped-def] self, data: Union[pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]]], group_by: str, return_df: bool = False, **calculate_kwargs, ) -> Union[List[pd.DataFrame], pd.DataFrame]: """Calculate features on each consecutive group of the data. Parameters ---------- data : Union[pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]]] Must be time-indexed! group_by: str Name of column by which to group values. return_df: bool, optional Whether the output needs to be a DataFrame or a list thereof, by default False. If `True` the output dataframes will be merged to a DataFrame with an outer merge. **calculate_kwargs: Keyword arguments that will be passed to the `calculate` method. .. Note:: Is comparable to following pseudo-SQL code: ```sql SELECT func(x) FROM `data` GROUP BY `group_by` ``` where `func` is the FeatureDescriptor function and `x` is the name on which the FeatureDescriptor operates. Note however that the grouping is done on consecutive values of `group_by` (i.e. `group_by` values that are the same and are next to each other are grouped together). """ # 0. Transform to dataframe series_dict = FeatureCollection._data_to_series_dict( data, self.get_required_series() + [group_by] ) df = pd.DataFrame(series_dict) # 1. Group by `group_by` column consecutive_grouped_by_df = self._group_by_consecutive(df, col_name=group_by) # 2. Get start and end idxs of consecutive groups start_segment_idxs = consecutive_grouped_by_df["start"] end_segment_idxs = start_segment_idxs.shift(-1) # fill the nan value with the last end idx end_segment_idxs.iloc[-1] = consecutive_grouped_by_df["end"].iloc[-1] # because segment end idxs are exclusive, we need to add an offset to the last # end idx so that all data gets used segment_vals = end_segment_idxs.values if is_datetime64_any_dtype(segment_vals): segment_vals[-1] += pd.Timedelta(days=1) else: segment_vals[-1] += 1 # 3. Calculate features try: # Filter out the warnings that are raised when segment indices are passed # (since users expect irregular window sizes when grouping) with warnings.catch_warnings(): warnings.filterwarnings( "ignore", category=RuntimeWarning, message="^.*segment indexes.*$" ) warnings.filterwarnings( "ignore", category=RuntimeWarning, message="^.*gaps.*$" ) # 3.1. Calculate features using the groupby segment idxs calc_results = self.calculate( data=df, segment_start_idxs=start_segment_idxs, segment_end_idxs=end_segment_idxs, **calculate_kwargs, ) # 3.2 Concatenate results and add the group_by column as well as the # start and end idxs of the segments calc_result = pd.concat(calc_results, join="outer", copy=False, axis=1) calc_result.reset_index(inplace=True, drop=True) calc_result[group_by] = consecutive_grouped_by_df[group_by] calc_result["__start"] = consecutive_grouped_by_df["start"] calc_result["__end"] = consecutive_grouped_by_df["end"] if return_df: return calc_result else: return [calc_result[col] for col in calc_result.columns] except Exception as e: raise RuntimeError( f"An exception was raised during feature extraction:\n{e}" ) @staticmethod def _process_njobs(n_jobs: Union[int, None], nb_funcs: int) -> int: """Process the number of jobs to run in parallel. On Windows no multiprocessing is supported, see https://github.com/predict-idlab/tsflex/issues/51 Parameters ---------- n_jobs : Union[int, None] The number of jobs to run in parallel. nb_funcs : int The number of feature functions. Returns ------- int The number of jobs to run in parallel. """ if os.name == "nt": # On Windows no multiprocessing is supported n_jobs = 1 else: n_jobs = parse_n_jobs(n_jobs) return min(n_jobs, nb_funcs) def _calculate_feature_list( self, executor: Callable[[int], pd.DataFrame], n_jobs: Union[int, None], show_progress: bool, return_df: bool, sort_output_index: bool, f_handler: Optional[logging.FileHandler], ) -> Union[List[pd.DataFrame], pd.DataFrame]: """Calculate the features for the given executor. Parameters ---------- executor : Callable[[int], pd.DataFrame] The executor function that will be used to calculate the features. n_jobs : Union[int, None] The number of jobs to run in parallel. show_progress : bool Whether to show a progress bar. return_df : bool Whether to return a DataFrame or a list of DataFrames. sort_output_index : bool Whether to sort the output index. Note that this is only relevant when `return_df` is set to `True`. f_handler : logging.FileHandler The file handler that is used to log the function execution times. Returns ------- Union[List[pd.DataFrame], pd.DataFrame] The calculated features. """ nb_feat_funcs = self._get_nb_feat_funcs() n_jobs = FeatureCollection._process_njobs(n_jobs, nb_feat_funcs) calculated_feature_list: Optional[List[pd.DataFrame]] = None if n_jobs in [0, 1]: # No multiprocessing idxs = range(nb_feat_funcs) if show_progress: idxs = tqdm(idxs) try: calculated_feature_list = [executor(idx) for idx in idxs] except Exception: traceback.print_exc() else: # Multiprocessing with Pool(processes=n_jobs) as pool: results = pool.imap_unordered(executor, range(nb_feat_funcs)) if show_progress: results = tqdm(results, total=nb_feat_funcs) try: calculated_feature_list = [f for f in results] except Exception: traceback.print_exc() pool.terminate() finally: # Close & join because: https://github.com/uqfoundation/pathos/issues/131 pool.close() pool.terminate() pool.join() # Close the file handler (this avoids PermissionError: [WinError 32]) if f_handler is not None: f_handler.close() logger.removeHandler(f_handler) if calculated_feature_list is None: raise RuntimeError( "Feature Extraction halted due to error while extracting one " + "(or multiple) feature(s)! See stack trace above." ) if return_df: # Concatenate & sort the columns df = pd.concat( calculated_feature_list, axis=1, join="outer", copy=False, sort=sort_output_index, ) return df.reindex(sorted(df.columns), axis=1) else: return calculated_feature_list def calculate( self, data: Union[ pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]], pd.core.groupby.DataFrameGroupby, ], stride: Optional[Union[float, str, pd.Timedelta, List, None]] = None, segment_start_idxs: Optional[ Union[list, np.ndarray, pd.Series, pd.Index] ] = None, segment_end_idxs: Optional[Union[list, np.ndarray, pd.Series, pd.Index]] = None, return_df: bool = False, window_idx: str = "end", include_final_window: bool = False, group_by_all: Optional[str] = None, # TODO: support multiple columns group_by_consecutive: Optional[str] = None, # TODO: support multiple columns bound_method: str = "inner", approve_sparsity: bool = False, show_progress: bool = False, logging_file_path: Optional[Union[str, Path]] = None, n_jobs: Optional[int] = None, ) -> Union[List[pd.DataFrame], pd.DataFrame]: """Calculate features on the passed data. Parameters ---------- data : Union[pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]], pd.core.groupby.DataFrameGroupby] Dataframe or Series or list thereof, with all the required data for the feature calculation. \n **Assumptions**: \n * each Series / DataFrame must have a sortable index. This index represents the sequence position of the corresponding values, the index can be either numeric or a ``pd.DatetimeIndex``. * each Series / DataFrame index must be comparable with all others * we assume that each series-name / dataframe-column-name is unique. Can also be a `DataFrameGroupBy` object, in which case the expected behaviour is similar to grouping by all values in `group_by_all` (i.e., for each group, the features are calculated on the group's data). stride: Union[float, str, pd.Timedelta, List[Union[float, str, pd.Timedelta], None], optional The stride size. By default None. This argument supports multiple types: \n * If None, the stride of the `FeatureDescriptor` objects will be used. * If the type is an `float` or an `int`, its value represents the series:\n - its stride **range** when a **non time-indexed** series is passed. - the stride in **number of samples**, when a **time-indexed** series is passed (must then be and `int`) * If the stride's type is a `pd.Timedelta`, the stride size represents the stride-time delta. The passed data **must have a time-index**. * If a `str`, it must represent a stride-time-delta-string. Hence, the **passed data must have a time-index**. \n .. Note:: When set, this stride argument takes precedence over the stride property of the `FeatureDescriptor`s in this `FeatureCollection` (i.e., when a not None value for `stride` passed to this method). segment_start_idxs: Union[list, np.ndarray, pd.Series, pd.Index], optional The start indices of the segments. If None, the start indices will be computed from the data using either:\n - the `segment_end_idxs` - the `window` size property of the `FeatureDescriptor` in this `FeatureCollection` (if `segment_end_idxs` is not None) - strided-window rolling on the data using `window` and `stride` of the `FeatureDescriptor` in this `FeatureCollection` (if `segment_end_idxs` is also None). (Note that the `stride` argument of this method takes precedence over the `stride` property of the `FeatureDescriptor`s). By default None. segment_end_idxs: Union[list, np.ndarray, pd.Series, pd.Index], optional The end indices for the segmented windows. If None, the end indices will be computed from the data using either:\n - the `segment_start_idxs` + the `window` size property of the `FeatureDescriptor` in this `FeatureCollection` (if `segment_start_idxs` is not None) - strided-window rolling on the data using `window` and `stride` of the `FeatureDescriptor` in this `FeatureCollection` (if `segment_start_idxs` is also None). (Note that the `stride` argument of this method takes precedence over the `stride` property of the `FeatureDescriptor`s). By default None. ..Note:: When passing both `segment_start_idxs` and `segment_end_idxs`, these two arguments must have the same length and every start index must be <= than the corresponding end index. Note that passing both arguments, discards any meaning of the `window` and `stride` values (as these segment indices define the segmented data, and thus no strided-window rolling index calculation has to be executed). As such, the user can create variable-length segmented windows. However, in such cases, the user should be weary that the feature functions are invariant to these (potentially variable-length) windows. return_df : bool, optional Whether the output needs to be a DataFrame or a list thereof, by default False. If `True` the output dataframes will be merged to a DataFrame with an outer merge. window_idx : str, optional The window's index position which will be used as index for the feature_window aggregation. Must be either of: `["begin", "middle", "end"]`. by **default "end"**. All features in this collection will use the same window_idx. ..Note:: `window_idx`="end" uses the window's end (= right open bound) as output index. \n `window_idx`="begin" uses the window's start idx (= left closed bound) as output index. include_final_window : bool, optional Whether the final (possibly incomplete) window should be included in the strided-window segmentation, by default False. .. Note:: The remarks below apply when `include_final_window` is set to True. The user should be aware that the last window *might* be incomplete, i.e.; - when equally sampled, the last window *might* be smaller than the the other windows. - when not equally sampled, the last window *might* not include all the data points (as the begin-time + window-size comes after the last data point). Note, that when equally sampled, the last window *will* be a full window when: - the stride is the sampling rate of the data (or stride = 1 for sample-based configurations).<br> **Remark**: that when `include_final_window` is set to False, the last window (which is a full) window will not be included! - *(len * sampling_rate - window_size) % stride = 0*. Remark that the above case is a base case of this. group_by_all : str, optional The name of the column by which to perform grouping. For each group, the features will be calculated. The output that is returned contains this `group_by` column as index to allow identifying the groups. If this parameter is used, the parameters `stride`, `segment_start_idxs`, `segment_end_idxs`, `window_idx` and `include_final_window` will be ignored. Rows with NaN values for this column will not be considered (as pandas ignores these rows when grouping). .. note:: This is similar as passing a `DataFrameGroupBy` object as `data` argument to the `calculate` method, where the `DataFrameGroupBy` object is created by calling `data.groupby(group_by_all)`. group_by_consecutive: str, optional The name of the column by which to perform consecutive grouping. A consecutive group is a group of values that are the same and are next to each other. For each consecutive group, the features will be calculated. The output that is returned contains this `group_by` column to allow identifying the groups, and also contains fields [`__start`, "__end"] which contain start and end time range for each result row. If this parameter is used, the parameters `stride`, `segment_start_idxs`, `segment_end_idxs`, `window_idx` and `include_final_window` will be ignored. Rows with NaN values for this column will not be considered (as we deem NaN not as a groupable value). Note that for consecutive grouping, groups can appear multiple times if they appear in different time-gaps. Example output: .. example:: ```python number_sold__sum__w=manual store __start __end 0 845 0 2019-01-01 2019-01-01 1 357 3 2019-01-02 2019-01-02 2 904 6 2019-01-03 2019-01-03 3 599 3 2019-01-04 2019-01-05 4 871 0 2019-01-06 2019-01-06 ... ... ... ... ... ``` bound_method: str, optional The start-end bound methodology which is used to generate the slice ranges when ``data`` consists of multiple series / columns. Must be either of: `["inner", "inner-outer", "outer"]`, by default "inner". * if ``inner``, the inner-bounds of the series are returned. * if ``inner-outer``, the left-inner and right-outer bounds of the series are returned. * if ``outer``, the outer-bounds of the series are returned. approve_sparsity: bool, optional Bool indicating whether the user acknowledges that there may be sparsity (i.e., irregularly sampled data), by default False. If False and sparsity is observed, a warning is raised. show_progress: bool, optional If True, the progress will be shown with a progressbar, by default False. logging_file_path : Union[str, Path], optional The file path where the logged messages are stored. If `None`, then no logging `FileHandler` will be used and the logging messages are only pushed to stdout. Otherwise, a logging `FileHandler` will write the logged messages to the given file path. See also the `tsflex.features.logger` module. n_jobs : int, optional The number of processes used for the feature calculation. If `None`, then the number returned by _os.cpu_count()_ is used, by default None. \n If n_jobs is either 0 or 1, the code will be executed sequentially without creating a process pool. This is very useful when debugging, as the stack trace will be more comprehensible. .. note:: Multiprocessed execution is not supported on Windows. Even when, `n_jobs` is set > 1, the feature extraction will still be executed sequentially. Why do we not support multiprocessing on Windows; see this issue https://github.com/predict-idlab/tsflex/issues/51 .. tip:: It takes on avg. _300ms_ to schedule everything with multiprocessing. So if your sequential feature extraction code runs faster than ~1s, it might not be worth it to parallelize the process (and thus better leave `n_jobs` to 0 or 1). Returns ------- Union[List[pd.DataFrame], pd.DataFrame] The calculated features. Raises ------ KeyError Raised when a required key is not found in `data`. Notes ------ * The (column-)names of the series in `data` represent the `series_names`. * If a `logging_file_path` is provided, the execution (time) info can be retrieved by calling `logger.get_feature_logs(logging_file_path)`. Be aware that the `logging_file_path` gets cleared before the logger pushes logged messages. Hence, one should use a separate logging file for each constructed processing and feature instance with this library. """ # Check valid data if isinstance(data, list): assert all( isinstance(d, (pd.Series, pd.DataFrame)) for d in data ), "All elements of the data list must be either a Series or a DataFrame!" else: assert isinstance( data, (pd.Series, pd.DataFrame, pd.core.groupby.DataFrameGroupBy) ), "The data must be either a Series, a DataFrame or a DataFrameGroupBy!" # check valid group_by assert group_by_all is None or group_by_consecutive is None, ( "Only max one of the following parameters can be set: " + "`group_by_all` or `group_by_consecutive`" ) assert not ( (group_by_all is not None or group_by_consecutive is not None) and isinstance(data, pd.core.groupby.DataFrameGroupBy) ), ( "Cannot use `group_by_all` or `group_by_consecutive` when `data` is" + " already a grouped DataFrame!" ) # Delete other logging handlers delete_logging_handlers(logger) # Add logging handler (if path provided) f_handler = None if logging_file_path: f_handler = add_logging_handler(logger, logging_file_path) if ( group_by_all or group_by_consecutive or isinstance(data, pd.core.groupby.DataFrameGroupBy) ): self._check_no_multiple_windows( error_case="When using the groupby behavior" ) # The grouping column must be part of the required series if group_by_all: # group_by_consecutive should be None (checked by asserts above) # data should not be a grouped DataFrame (checked by asserts above) assert ( group_by_all not in self.get_required_series() ), "The `group_by_all` column cannot be part of the required series!" elif group_by_consecutive: # group_by_all should be None (checked by asserts above) # data should not be a grouped DataFrame (checked by asserts above) assert group_by_consecutive not in self.get_required_series(), ( "The `group_by_consecutive` column cannot be part of the required " + "series!" ) # __start and __end should not be part of the output names assert "__start" not in self.get_required_series() assert "__end" not in self.get_required_series() # if any of the following params are not None, warn that they won't be of use # in the grouped calculation ignored_params = [ ("stride", None), ("segment_start_idxs", None), ("segment_end_idxs", None), ("window_idx", "end"), ("include_final_window", False), ] local_params = locals() for ip, default_value in ignored_params: if local_params[ip] is not default_value: warnings.warn( f"Parameter `{ip}` will be ignored in case of GroupBy feature" + " calculation." ) if group_by_consecutive: # Strided rollling feature extraction will take place return self._calculate_group_by_consecutive( data, group_by_consecutive, return_df, bound_method=bound_method, approve_sparsity=approve_sparsity, show_progress=show_progress, logging_file_path=logging_file_path, n_jobs=n_jobs, ) else: # Grouped feature extraction will take place if not isinstance(data, pd.core.groupby.generic.DataFrameGroupBy): # group_by_all should not be None (checked by asserts above) assert group_by_all is not None # 0. Transform to dataframe series_dict = FeatureCollection._data_to_series_dict( data, self.get_required_series() + [group_by_all] ) # 1. Group by `group_by_all` column data = self._group_by_all(series_dict, col_name=group_by_all) return self._calculate_group_by_all( data, # should be a DataFrameGroupBy return_df, show_progress=show_progress, n_jobs=n_jobs, f_handler=f_handler, ) # Sort output index if segment indices are not provided sort_output_index = segment_start_idxs is None and segment_end_idxs is None # Convert to numpy array (if necessary) if segment_start_idxs is not None: segment_start_idxs = FeatureCollection._process_segment_idxs( segment_start_idxs ) if segment_end_idxs is not None: segment_end_idxs = FeatureCollection._process_segment_idxs(segment_end_idxs) if segment_start_idxs is not None and segment_end_idxs is not None: # Check if segment indices have same length and whether every start idx # <= end idx _check_start_end_array(segment_start_idxs, segment_end_idxs) # Check if there is either 1 or No(ne) window value for every output name - # input_series combination self._check_no_multiple_windows( error_case="When using both `segment_start_idxs` and `segment_end_idxs`" ) if segment_start_idxs is None or segment_end_idxs is None: assert all( fd.window is not None for fd in flatten(self._feature_desc_dict.values()) ), ( "Each feature descriptor must have a window when not both " + "segment_start_idxs and segment_end_idxs are provided" ) if stride is None and segment_start_idxs is None and segment_end_idxs is None: assert all( fd.stride is not None for fd in flatten(self._feature_desc_dict.values()) ), ( "Each feature descriptor must have a stride when no stride or " + "segment indices are passed to this method!" ) elif stride is not None and ( segment_start_idxs is not None or segment_end_idxs is not None ): raise ValueError( "The stride and any segment index argument cannot be set together! " + "At least one of both should be None." ) if stride is not None: # Verify whether the stride complies with the input data dtype stride = [ parse_time_arg(s) if isinstance(s, str) else s for s in to_list(stride) ] self._check_feature_descriptors(skip_none=False, calc_stride=stride) # Convert the data to a series_dict series_dict = FeatureCollection._data_to_series_dict( data, self.get_required_series() ) # Determine the bounds of the series dict items and slice on them # TODO: is dit wel nodig `hier? want we doen dat ook in de strided rolling start, end = _determine_bounds(bound_method, list(series_dict.values())) series_dict = { n: s.loc[ s.index.dtype.type(start) : s.index.dtype.type(end) ] # TODO: check memory efficiency of ths for n, s, in series_dict.items() } # Note: this variable has a global scope so this is shared in multiprocessing # TODO: try to make this more efficient (but is not really the bottleneck) global get_stroll_func get_stroll_func = self._stroll_feat_generator( series_dict, calc_stride=stride, segment_start_idxs=segment_start_idxs, segment_end_idxs=segment_end_idxs, start_idx=start, end_idx=end, window_idx=window_idx, include_final_window=include_final_window, approve_sparsity=approve_sparsity, ) return self._calculate_feature_list( self._executor_stroll, n_jobs, show_progress, return_df, sort_output_index, f_handler, ) def serialize(self, file_path: Union[str, Path]) -> None: """Serialize this FeatureCollection instance. Parameters ---------- file_path : Union[str, Path] The path where the `FeatureCollection` will be serialized. Note ----- As we use [Dill](https://github.com/uqfoundation/dill){:target="_blank"} to serialize the files, we can **also serialize functions which are defined in the local scope, like lambdas.** """ with open(file_path, "wb") as f: dill.dump(self, f, recurse=True) def reduce(self, feat_cols_to_keep: List[str]) -> FeatureCollection: """Create a reduced FeatureCollection instance based on `feat_cols_to_keep`. For example, this is useful to optimize feature-extraction inference (for your selected features) after performing a feature-selection procedure. Parameters ---------- feat_cols_to_keep: List[str] A subset of the feature collection instance its column names. This corresponds to the columns / names of the output from `calculate` method that you want to keep. Returns ------- FeatureCollection A new FeatureCollection object, which only withholds the FeatureDescriptors which constitute the `feat_cols_to_keep` output. Note ---- Some FeatureDescriptor objects may have multiple **output-names**.<br> Hence, if you only want to retain _a subset_ of that FeatureDescriptor its feature outputs, you will still get **all features** as the new FeatureCollection is constructed by applying a filter on de FeatureDescriptor list and we thus not alter these FeatureDescriptor objects themselves. """ # dict in which we store all the { output_col_name : (UUID, FeatureDescriptor) } # items of our current FeatureCollection object manual_window = False if any(c.endswith("w=manual") for c in feat_cols_to_keep): assert all(c.endswith("w=manual") for c in feat_cols_to_keep) # As the windows are created manual, the FeatureCollection cannot contain # multiple windows for the same output name - input_series combination self._check_no_multiple_windows( error_case="When reducing a FeatureCollection with manual windows" ) manual_window = True feat_col_fd_mapping: Dict[str, Tuple[str, FeatureDescriptor]] = {} for (s_names, window), fd_list in self._feature_desc_dict.items(): window = "manual" if manual_window else self._ws_to_str(window) for fd in fd_list: # As a single FeatureDescriptor can have multiple output col names, we # create a unique identifier for each FeatureDescriptor (on which we # will apply set-like operations later on to only retain all the unique # FeatureDescriptors) uuid_str = str(uuid.uuid4()) for output_name in fd.function.output_names: # Reconstruct the feature column name feat_col_name = StridedRolling.construct_output_index( series_keys=s_names, feat_name=output_name, win_str=window ) feat_col_fd_mapping[feat_col_name] = (uuid_str, fd) assert all(fc in feat_col_fd_mapping for fc in feat_cols_to_keep) # Collect (uuid, FeatureDescriptor) for the feat_cols_to_keep fd_subset: List[Tuple[str, FeatureDescriptor]] = [ feat_col_fd_mapping[fc] for fc in feat_cols_to_keep ] # Reduce to unique feature descriptor objects (based on uuid) and create a new # FeatureCollection for their deepcopy's. seen_uuids = set() fds = [] for uuid_str, fd in fd_subset: if uuid_str not in seen_uuids: seen_uuids.add(uuid_str) fds.append(deepcopy(fd)) return FeatureCollection(feature_descriptors=fds) @staticmethod def _ws_to_str(window_or_stride: Any) -> str: """Convert the window/stride value to a (shortend) string representation.""" if isinstance(window_or_stride, pd.Timedelta): return timedelta_to_str(window_or_stride) else: return str(window_or_stride) def __repr__(self) -> str: """Representation string of a FeatureCollection.""" feature_keys = sorted(set(k[0] for k in self._feature_desc_dict.keys())) output_str = "" for feature_key in feature_keys: output_str += f"{'|'.join(feature_key)}: (" keys = (x for x in self._feature_desc_dict.keys() if x[0] == feature_key) for _, win_size in keys: output_str += "\n\twin: " win_str = self._ws_to_str(win_size) output_str += f"{win_str:<6}: [" for feat_desc in self._feature_desc_dict[feature_key, win_size]: stride_str = feat_desc.stride if stride_str is not None: stride_str = [self._ws_to_str(s) for s in stride_str] output_str += f"\n\t\t{feat_desc._func_str}" output_str += f" stride: {stride_str}," output_str += "\n\t]" output_str += "\n)\n" return output_strCreate a FeatureCollection.
Parameters
feature_descriptors:Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection, List[Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection]]], optional- Initial (list of) feature(s) to add to collection, by default None
Notes
- The
series_nameproperty of theFeatureDescriptors should not withhold a "|" character, since "|" is used to join the series names of features which use multiple series as input).
e.g.ACC|xis not allowed as series name, as this is ambiguous and could represent that this feature is constructed with a combination of theACCandxsignal.
Note thatmax|featis allowed as feature output name.
- Both the
series_nameandoutput_nameproperty of theFeatureDescriptors should not withhold "__" in its string representations. This constraint is mainly made for readability purposes.
The two statements above will be asserted
Methods
def get_required_series(self)-
Expand source code
def get_required_series(self) -> List[str]: """Return all required series names for this feature collection. Return the list of series names that are required in order to calculate all the features (defined by the `FeatureDescriptor` objects) of this feature collection. Returns ------- List[str] List of all the required series names. """ return list( set(flatten([fr_key[0] for fr_key in self._feature_desc_dict.keys()])) )Return all required series names for this feature collection.
Return the list of series names that are required in order to calculate all the features (defined by the
FeatureDescriptorobjects) of this feature collection.Returns
List[str]- List of all the required series names.
def get_nb_output_features(self)-
Expand source code
def get_nb_output_features(self) -> int: """Return the number of output features in this feature collection. Returns ------- int The number of output features in this feature collection. """ fd_list: Iterable[FeatureDescriptor] = flatten(self._feature_desc_dict.values()) return sum(fd.get_nb_output_features() for fd in fd_list)Return the number of output features in this feature collection.
Returns
int- The number of output features in this feature collection.
def add(self, features)-
Expand source code
def add( self, features: Union[ FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection, Sequence[ Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection] ], ], ) -> None: """Add feature(s) to the FeatureCollection. Parameters ---------- features : Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection, List[Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection]]] Feature(s) (containers) whose contained features will be added. Raises ------ TypeError Raised when an item within `features` is not an instance of [`MultipleFeatureDescriptors`, `FeatureDescriptors`, `FeatureCollection`]. """ # Convert to list if necessary features = to_list(features) for feature in features: if isinstance(feature, MultipleFeatureDescriptors): self.add(feature.feature_descriptions) elif isinstance(feature, FeatureDescriptor): self._add_feature(feature) elif isinstance(feature, FeatureCollection): # List needs to be flattened self.add(list(flatten(feature._feature_desc_dict.values()))) else: raise TypeError(f"type: {type(feature)} is not supported - {feature}") # After adding the features, check whether the descriptors are compatible self._check_feature_descriptors(skip_none=True)Add feature(s) to the FeatureCollection.
Parameters
features:Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection, List[Union[FeatureDescriptor, MultipleFeatureDescriptors, FeatureCollection]]]- Feature(s) (containers) whose contained features will be added.
Raises
TypeError- Raised when an item within
featuresis not an instance of [MultipleFeatureDescriptors,FeatureDescriptors,FeatureCollection].
def calculate(self, data, stride=None, segment_start_idxs=None, segment_end_idxs=None, return_df=False, window_idx='end', include_final_window=False, group_by_all=None, group_by_consecutive=None, bound_method='inner', approve_sparsity=False, show_progress=False, logging_file_path=None, n_jobs=None)-
Expand source code
def calculate( self, data: Union[ pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]], pd.core.groupby.DataFrameGroupby, ], stride: Optional[Union[float, str, pd.Timedelta, List, None]] = None, segment_start_idxs: Optional[ Union[list, np.ndarray, pd.Series, pd.Index] ] = None, segment_end_idxs: Optional[Union[list, np.ndarray, pd.Series, pd.Index]] = None, return_df: bool = False, window_idx: str = "end", include_final_window: bool = False, group_by_all: Optional[str] = None, # TODO: support multiple columns group_by_consecutive: Optional[str] = None, # TODO: support multiple columns bound_method: str = "inner", approve_sparsity: bool = False, show_progress: bool = False, logging_file_path: Optional[Union[str, Path]] = None, n_jobs: Optional[int] = None, ) -> Union[List[pd.DataFrame], pd.DataFrame]: """Calculate features on the passed data. Parameters ---------- data : Union[pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]], pd.core.groupby.DataFrameGroupby] Dataframe or Series or list thereof, with all the required data for the feature calculation. \n **Assumptions**: \n * each Series / DataFrame must have a sortable index. This index represents the sequence position of the corresponding values, the index can be either numeric or a ``pd.DatetimeIndex``. * each Series / DataFrame index must be comparable with all others * we assume that each series-name / dataframe-column-name is unique. Can also be a `DataFrameGroupBy` object, in which case the expected behaviour is similar to grouping by all values in `group_by_all` (i.e., for each group, the features are calculated on the group's data). stride: Union[float, str, pd.Timedelta, List[Union[float, str, pd.Timedelta], None], optional The stride size. By default None. This argument supports multiple types: \n * If None, the stride of the `FeatureDescriptor` objects will be used. * If the type is an `float` or an `int`, its value represents the series:\n - its stride **range** when a **non time-indexed** series is passed. - the stride in **number of samples**, when a **time-indexed** series is passed (must then be and `int`) * If the stride's type is a `pd.Timedelta`, the stride size represents the stride-time delta. The passed data **must have a time-index**. * If a `str`, it must represent a stride-time-delta-string. Hence, the **passed data must have a time-index**. \n .. Note:: When set, this stride argument takes precedence over the stride property of the `FeatureDescriptor`s in this `FeatureCollection` (i.e., when a not None value for `stride` passed to this method). segment_start_idxs: Union[list, np.ndarray, pd.Series, pd.Index], optional The start indices of the segments. If None, the start indices will be computed from the data using either:\n - the `segment_end_idxs` - the `window` size property of the `FeatureDescriptor` in this `FeatureCollection` (if `segment_end_idxs` is not None) - strided-window rolling on the data using `window` and `stride` of the `FeatureDescriptor` in this `FeatureCollection` (if `segment_end_idxs` is also None). (Note that the `stride` argument of this method takes precedence over the `stride` property of the `FeatureDescriptor`s). By default None. segment_end_idxs: Union[list, np.ndarray, pd.Series, pd.Index], optional The end indices for the segmented windows. If None, the end indices will be computed from the data using either:\n - the `segment_start_idxs` + the `window` size property of the `FeatureDescriptor` in this `FeatureCollection` (if `segment_start_idxs` is not None) - strided-window rolling on the data using `window` and `stride` of the `FeatureDescriptor` in this `FeatureCollection` (if `segment_start_idxs` is also None). (Note that the `stride` argument of this method takes precedence over the `stride` property of the `FeatureDescriptor`s). By default None. ..Note:: When passing both `segment_start_idxs` and `segment_end_idxs`, these two arguments must have the same length and every start index must be <= than the corresponding end index. Note that passing both arguments, discards any meaning of the `window` and `stride` values (as these segment indices define the segmented data, and thus no strided-window rolling index calculation has to be executed). As such, the user can create variable-length segmented windows. However, in such cases, the user should be weary that the feature functions are invariant to these (potentially variable-length) windows. return_df : bool, optional Whether the output needs to be a DataFrame or a list thereof, by default False. If `True` the output dataframes will be merged to a DataFrame with an outer merge. window_idx : str, optional The window's index position which will be used as index for the feature_window aggregation. Must be either of: `["begin", "middle", "end"]`. by **default "end"**. All features in this collection will use the same window_idx. ..Note:: `window_idx`="end" uses the window's end (= right open bound) as output index. \n `window_idx`="begin" uses the window's start idx (= left closed bound) as output index. include_final_window : bool, optional Whether the final (possibly incomplete) window should be included in the strided-window segmentation, by default False. .. Note:: The remarks below apply when `include_final_window` is set to True. The user should be aware that the last window *might* be incomplete, i.e.; - when equally sampled, the last window *might* be smaller than the the other windows. - when not equally sampled, the last window *might* not include all the data points (as the begin-time + window-size comes after the last data point). Note, that when equally sampled, the last window *will* be a full window when: - the stride is the sampling rate of the data (or stride = 1 for sample-based configurations).<br> **Remark**: that when `include_final_window` is set to False, the last window (which is a full) window will not be included! - *(len * sampling_rate - window_size) % stride = 0*. Remark that the above case is a base case of this. group_by_all : str, optional The name of the column by which to perform grouping. For each group, the features will be calculated. The output that is returned contains this `group_by` column as index to allow identifying the groups. If this parameter is used, the parameters `stride`, `segment_start_idxs`, `segment_end_idxs`, `window_idx` and `include_final_window` will be ignored. Rows with NaN values for this column will not be considered (as pandas ignores these rows when grouping). .. note:: This is similar as passing a `DataFrameGroupBy` object as `data` argument to the `calculate` method, where the `DataFrameGroupBy` object is created by calling `data.groupby(group_by_all)`. group_by_consecutive: str, optional The name of the column by which to perform consecutive grouping. A consecutive group is a group of values that are the same and are next to each other. For each consecutive group, the features will be calculated. The output that is returned contains this `group_by` column to allow identifying the groups, and also contains fields [`__start`, "__end"] which contain start and end time range for each result row. If this parameter is used, the parameters `stride`, `segment_start_idxs`, `segment_end_idxs`, `window_idx` and `include_final_window` will be ignored. Rows with NaN values for this column will not be considered (as we deem NaN not as a groupable value). Note that for consecutive grouping, groups can appear multiple times if they appear in different time-gaps. Example output: .. example:: ```python number_sold__sum__w=manual store __start __end 0 845 0 2019-01-01 2019-01-01 1 357 3 2019-01-02 2019-01-02 2 904 6 2019-01-03 2019-01-03 3 599 3 2019-01-04 2019-01-05 4 871 0 2019-01-06 2019-01-06 ... ... ... ... ... ``` bound_method: str, optional The start-end bound methodology which is used to generate the slice ranges when ``data`` consists of multiple series / columns. Must be either of: `["inner", "inner-outer", "outer"]`, by default "inner". * if ``inner``, the inner-bounds of the series are returned. * if ``inner-outer``, the left-inner and right-outer bounds of the series are returned. * if ``outer``, the outer-bounds of the series are returned. approve_sparsity: bool, optional Bool indicating whether the user acknowledges that there may be sparsity (i.e., irregularly sampled data), by default False. If False and sparsity is observed, a warning is raised. show_progress: bool, optional If True, the progress will be shown with a progressbar, by default False. logging_file_path : Union[str, Path], optional The file path where the logged messages are stored. If `None`, then no logging `FileHandler` will be used and the logging messages are only pushed to stdout. Otherwise, a logging `FileHandler` will write the logged messages to the given file path. See also the `tsflex.features.logger` module. n_jobs : int, optional The number of processes used for the feature calculation. If `None`, then the number returned by _os.cpu_count()_ is used, by default None. \n If n_jobs is either 0 or 1, the code will be executed sequentially without creating a process pool. This is very useful when debugging, as the stack trace will be more comprehensible. .. note:: Multiprocessed execution is not supported on Windows. Even when, `n_jobs` is set > 1, the feature extraction will still be executed sequentially. Why do we not support multiprocessing on Windows; see this issue https://github.com/predict-idlab/tsflex/issues/51 .. tip:: It takes on avg. _300ms_ to schedule everything with multiprocessing. So if your sequential feature extraction code runs faster than ~1s, it might not be worth it to parallelize the process (and thus better leave `n_jobs` to 0 or 1). Returns ------- Union[List[pd.DataFrame], pd.DataFrame] The calculated features. Raises ------ KeyError Raised when a required key is not found in `data`. Notes ------ * The (column-)names of the series in `data` represent the `series_names`. * If a `logging_file_path` is provided, the execution (time) info can be retrieved by calling `logger.get_feature_logs(logging_file_path)`. Be aware that the `logging_file_path` gets cleared before the logger pushes logged messages. Hence, one should use a separate logging file for each constructed processing and feature instance with this library. """ # Check valid data if isinstance(data, list): assert all( isinstance(d, (pd.Series, pd.DataFrame)) for d in data ), "All elements of the data list must be either a Series or a DataFrame!" else: assert isinstance( data, (pd.Series, pd.DataFrame, pd.core.groupby.DataFrameGroupBy) ), "The data must be either a Series, a DataFrame or a DataFrameGroupBy!" # check valid group_by assert group_by_all is None or group_by_consecutive is None, ( "Only max one of the following parameters can be set: " + "`group_by_all` or `group_by_consecutive`" ) assert not ( (group_by_all is not None or group_by_consecutive is not None) and isinstance(data, pd.core.groupby.DataFrameGroupBy) ), ( "Cannot use `group_by_all` or `group_by_consecutive` when `data` is" + " already a grouped DataFrame!" ) # Delete other logging handlers delete_logging_handlers(logger) # Add logging handler (if path provided) f_handler = None if logging_file_path: f_handler = add_logging_handler(logger, logging_file_path) if ( group_by_all or group_by_consecutive or isinstance(data, pd.core.groupby.DataFrameGroupBy) ): self._check_no_multiple_windows( error_case="When using the groupby behavior" ) # The grouping column must be part of the required series if group_by_all: # group_by_consecutive should be None (checked by asserts above) # data should not be a grouped DataFrame (checked by asserts above) assert ( group_by_all not in self.get_required_series() ), "The `group_by_all` column cannot be part of the required series!" elif group_by_consecutive: # group_by_all should be None (checked by asserts above) # data should not be a grouped DataFrame (checked by asserts above) assert group_by_consecutive not in self.get_required_series(), ( "The `group_by_consecutive` column cannot be part of the required " + "series!" ) # __start and __end should not be part of the output names assert "__start" not in self.get_required_series() assert "__end" not in self.get_required_series() # if any of the following params are not None, warn that they won't be of use # in the grouped calculation ignored_params = [ ("stride", None), ("segment_start_idxs", None), ("segment_end_idxs", None), ("window_idx", "end"), ("include_final_window", False), ] local_params = locals() for ip, default_value in ignored_params: if local_params[ip] is not default_value: warnings.warn( f"Parameter `{ip}` will be ignored in case of GroupBy feature" + " calculation." ) if group_by_consecutive: # Strided rollling feature extraction will take place return self._calculate_group_by_consecutive( data, group_by_consecutive, return_df, bound_method=bound_method, approve_sparsity=approve_sparsity, show_progress=show_progress, logging_file_path=logging_file_path, n_jobs=n_jobs, ) else: # Grouped feature extraction will take place if not isinstance(data, pd.core.groupby.generic.DataFrameGroupBy): # group_by_all should not be None (checked by asserts above) assert group_by_all is not None # 0. Transform to dataframe series_dict = FeatureCollection._data_to_series_dict( data, self.get_required_series() + [group_by_all] ) # 1. Group by `group_by_all` column data = self._group_by_all(series_dict, col_name=group_by_all) return self._calculate_group_by_all( data, # should be a DataFrameGroupBy return_df, show_progress=show_progress, n_jobs=n_jobs, f_handler=f_handler, ) # Sort output index if segment indices are not provided sort_output_index = segment_start_idxs is None and segment_end_idxs is None # Convert to numpy array (if necessary) if segment_start_idxs is not None: segment_start_idxs = FeatureCollection._process_segment_idxs( segment_start_idxs ) if segment_end_idxs is not None: segment_end_idxs = FeatureCollection._process_segment_idxs(segment_end_idxs) if segment_start_idxs is not None and segment_end_idxs is not None: # Check if segment indices have same length and whether every start idx # <= end idx _check_start_end_array(segment_start_idxs, segment_end_idxs) # Check if there is either 1 or No(ne) window value for every output name - # input_series combination self._check_no_multiple_windows( error_case="When using both `segment_start_idxs` and `segment_end_idxs`" ) if segment_start_idxs is None or segment_end_idxs is None: assert all( fd.window is not None for fd in flatten(self._feature_desc_dict.values()) ), ( "Each feature descriptor must have a window when not both " + "segment_start_idxs and segment_end_idxs are provided" ) if stride is None and segment_start_idxs is None and segment_end_idxs is None: assert all( fd.stride is not None for fd in flatten(self._feature_desc_dict.values()) ), ( "Each feature descriptor must have a stride when no stride or " + "segment indices are passed to this method!" ) elif stride is not None and ( segment_start_idxs is not None or segment_end_idxs is not None ): raise ValueError( "The stride and any segment index argument cannot be set together! " + "At least one of both should be None." ) if stride is not None: # Verify whether the stride complies with the input data dtype stride = [ parse_time_arg(s) if isinstance(s, str) else s for s in to_list(stride) ] self._check_feature_descriptors(skip_none=False, calc_stride=stride) # Convert the data to a series_dict series_dict = FeatureCollection._data_to_series_dict( data, self.get_required_series() ) # Determine the bounds of the series dict items and slice on them # TODO: is dit wel nodig `hier? want we doen dat ook in de strided rolling start, end = _determine_bounds(bound_method, list(series_dict.values())) series_dict = { n: s.loc[ s.index.dtype.type(start) : s.index.dtype.type(end) ] # TODO: check memory efficiency of ths for n, s, in series_dict.items() } # Note: this variable has a global scope so this is shared in multiprocessing # TODO: try to make this more efficient (but is not really the bottleneck) global get_stroll_func get_stroll_func = self._stroll_feat_generator( series_dict, calc_stride=stride, segment_start_idxs=segment_start_idxs, segment_end_idxs=segment_end_idxs, start_idx=start, end_idx=end, window_idx=window_idx, include_final_window=include_final_window, approve_sparsity=approve_sparsity, ) return self._calculate_feature_list( self._executor_stroll, n_jobs, show_progress, return_df, sort_output_index, f_handler, )Calculate features on the passed data.
Parameters
data:Union[pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]], pd.core.groupby.DataFrameGroupby]-
Dataframe or Series or list thereof, with all the required data for the feature calculation.
Assumptions:
- each Series / DataFrame must have a sortable index. This index represents
the sequence position of the corresponding values, the index can be either
numeric or a
pd.DatetimeIndex. - each Series / DataFrame index must be comparable with all others
- we assume that each series-name / dataframe-column-name is unique.
Can also be a
DataFrameGroupByobject, in which case the expected behaviour is similar to grouping by all values ingroup_by_all(i.e., for each group, the features are calculated on the group's data).
- each Series / DataFrame must have a sortable index. This index represents
the sequence position of the corresponding values, the index can be either
numeric or a
stride:Union[float, str, pd.Timedelta, List[Union[float, str, pd.Timedelta], None], optional-
The stride size. By default None. This argument supports multiple types:
- If None, the stride of the
FeatureDescriptorobjects will be used. -
If the type is an
floator anint, its value represents the series:- its stride range when a non time-indexed series is passed.
- the stride in number of samples, when a time-indexed series
is passed (must then be and
int)- If the stride's type is a
pd.Timedelta, the stride size represents the stride-time delta. The passed data must have a time-index. - If a
str, it must represent a stride-time-delta-string. Hence, the passed data must have a time-index.
- If the stride's type is a
Note
When set, this stride argument takes precedence over the stride property of the
FeatureDescriptors in thisFeatureCollection(i.e., when a not None value forstridepassed to this method). - If None, the stride of the
segment_start_idxs:Union[list, np.ndarray, pd.Series, pd.Index], optional-
The start indices of the segments. If None, the start indices will be computed from the data using either:
- the
segment_end_idxs- thewindowsize property of theFeatureDescriptorin thisFeatureCollection(ifsegment_end_idxsis not None) - strided-window rolling on the data using
windowandstrideof theFeatureDescriptorin thisFeatureCollection(ifsegment_end_idxsis also None). (Note that thestrideargument of this method takes precedence over thestrideproperty of theFeatureDescriptors). By default None.
- the
segment_end_idxs:Union[list, np.ndarray, pd.Series, pd.Index], optional-
The end indices for the segmented windows. If None, the end indices will be computed from the data using either:
- the
segment_start_idxs+ thewindowsize property of theFeatureDescriptorin thisFeatureCollection(ifsegment_start_idxsis not None) - strided-window rolling on the data using
windowandstrideof theFeatureDescriptorin thisFeatureCollection(ifsegment_start_idxsis also None). (Note that thestrideargument of this method takes precedence over thestrideproperty of theFeatureDescriptors). By default None.
Note
When passing both
segment_start_idxsandsegment_end_idxs, these two arguments must have the same length and every start index must be <= than the corresponding end index. Note that passing both arguments, discards any meaning of thewindowandstridevalues (as these segment indices define the segmented data, and thus no strided-window rolling index calculation has to be executed). As such, the user can create variable-length segmented windows. However, in such cases, the user should be weary that the feature functions are invariant to these (potentially variable-length) windows. - the
return_df:bool, optional- Whether the output needs to be a DataFrame or a list thereof, by default
False. If
Truethe output dataframes will be merged to a DataFrame with an outer merge. window_idx:str, optional-
The window's index position which will be used as index for the feature_window aggregation. Must be either of:
["begin", "middle", "end"]. by default "end". All features in this collection will use the same window_idx.Note
window_idx="end" uses the window's end (= right open bound) as output index.window_idx="begin" uses the window's start idx (= left closed bound) as output index. include_final_window:bool, optional-
Whether the final (possibly incomplete) window should be included in the strided-window segmentation, by default False.
Note
The remarks below apply when
include_final_windowis set to True. The user should be aware that the last window might be incomplete, i.e.;- when equally sampled, the last window might be smaller than the the other windows.
- when not equally sampled, the last window might not include all the data points (as the begin-time + window-size comes after the last data point).
Note, that when equally sampled, the last window will be a full window when:
- the stride is the sampling rate of the data (or stride = 1 for
sample-based configurations).
Remark: that wheninclude_final_windowis set to False, the last window (which is a full) window will not be included! - (len * sampling_rate - window_size) % stride = 0. Remark that the above case is a base case of this.
group_by_all:str, optional- The name of the column by which to perform grouping. For each group, the
features will be calculated. The output that is returned contains this
group_bycolumn as index to allow identifying the groups. If this parameter is used, the parametersstride,segment_start_idxs,segment_end_idxs,window_idxandinclude_final_windowwill be ignored. Rows with NaN values for this column will not be considered (as pandas ignores these rows when grouping).Note
This is similar as passing aDataFrameGroupByobject asdataargument to thecalculatemethod, where theDataFrameGroupByobject is created by callingdata.groupby(group_by_all). group_by_consecutive:str, optional-
The name of the column by which to perform consecutive grouping. A consecutive group is a group of values that are the same and are next to each other. For each consecutive group, the features will be calculated. The output that is returned contains this
group_bycolumn to allow identifying the groups, and also contains fields [__start, "__end"] which contain start and end time range for each result row. If this parameter is used, the parametersstride,segment_start_idxs,segment_end_idxs,window_idxandinclude_final_windowwill be ignored. Rows with NaN values for this column will not be considered (as we deem NaN not as a groupable value). Note that for consecutive grouping, groups can appear multiple times if they appear in different time-gaps.Example output:
Example
number_sold__sum__w=manual store __start __end 0 845 0 2019-01-01 2019-01-01 1 357 3 2019-01-02 2019-01-02 2 904 6 2019-01-03 2019-01-03 3 599 3 2019-01-04 2019-01-05 4 871 0 2019-01-06 2019-01-06 ... ... ... ... ...bound_method:str, optional-
The start-end bound methodology which is used to generate the slice ranges when
dataconsists of multiple series / columns. Must be either of:["inner", "inner-outer", "outer"], by default "inner".- if
inner, the inner-bounds of the series are returned. - if
inner-outer, the left-inner and right-outer bounds of the series are returned. - if
outer, the outer-bounds of the series are returned.
- if
approve_sparsity:bool, optional- Bool indicating whether the user acknowledges that there may be sparsity (i.e., irregularly sampled data), by default False. If False and sparsity is observed, a warning is raised.
show_progress:bool, optional- If True, the progress will be shown with a progressbar, by default False.
logging_file_path:Union[str, Path], optional- The file path where the logged messages are stored. If
None, then no loggingFileHandlerwill be used and the logging messages are only pushed to stdout. Otherwise, a loggingFileHandlerwill write the logged messages to the given file path. See also the.loggermodule. n_jobs:int, optional-
The number of processes used for the feature calculation. If
None, then the number returned by os.cpu_count() is used, by default None.If n_jobs is either 0 or 1, the code will be executed sequentially without creating a process pool. This is very useful when debugging, as the stack trace will be more comprehensible.
Note
Multiprocessed execution is not supported on Windows. Even when,
n_jobsis set > 1, the feature extraction will still be executed sequentially. Why do we not support multiprocessing on Windows; see this issue https://github.com/predict-idlab/tsflex/issues/51Tip
It takes on avg. 300ms to schedule everything with multiprocessing. So if your sequential feature extraction code runs faster than ~1s, it might not be worth it to parallelize the process (and thus better leave
n_jobsto 0 or 1).
Returns
Union[List[pd.DataFrame], pd.DataFrame]- The calculated features.
Raises
KeyError- Raised when a required key is not found in
data.
Notes
- The (column-)names of the series in
datarepresent theseries_names. - If a
logging_file_pathis provided, the execution (time) info can be retrieved by callingget_feature_logs()(logging_file_path). Be aware that thelogging_file_pathgets cleared before the logger pushes logged messages. Hence, one should use a separate logging file for each constructed processing and feature instance with this library.
def serialize(self, file_path)-
Expand source code
def serialize(self, file_path: Union[str, Path]) -> None: """Serialize this FeatureCollection instance. Parameters ---------- file_path : Union[str, Path] The path where the `FeatureCollection` will be serialized. Note ----- As we use [Dill](https://github.com/uqfoundation/dill){:target="_blank"} to serialize the files, we can **also serialize functions which are defined in the local scope, like lambdas.** """ with open(file_path, "wb") as f: dill.dump(self, f, recurse=True)Serialize this FeatureCollection instance.
Parameters
file_path:Union[str, Path]- The path where the
FeatureCollectionwill be serialized.
Note
As we use Dill to serialize the files, we can also serialize functions which are defined in the local scope, like lambdas.
def reduce(self, feat_cols_to_keep)-
Expand source code
def reduce(self, feat_cols_to_keep: List[str]) -> FeatureCollection: """Create a reduced FeatureCollection instance based on `feat_cols_to_keep`. For example, this is useful to optimize feature-extraction inference (for your selected features) after performing a feature-selection procedure. Parameters ---------- feat_cols_to_keep: List[str] A subset of the feature collection instance its column names. This corresponds to the columns / names of the output from `calculate` method that you want to keep. Returns ------- FeatureCollection A new FeatureCollection object, which only withholds the FeatureDescriptors which constitute the `feat_cols_to_keep` output. Note ---- Some FeatureDescriptor objects may have multiple **output-names**.<br> Hence, if you only want to retain _a subset_ of that FeatureDescriptor its feature outputs, you will still get **all features** as the new FeatureCollection is constructed by applying a filter on de FeatureDescriptor list and we thus not alter these FeatureDescriptor objects themselves. """ # dict in which we store all the { output_col_name : (UUID, FeatureDescriptor) } # items of our current FeatureCollection object manual_window = False if any(c.endswith("w=manual") for c in feat_cols_to_keep): assert all(c.endswith("w=manual") for c in feat_cols_to_keep) # As the windows are created manual, the FeatureCollection cannot contain # multiple windows for the same output name - input_series combination self._check_no_multiple_windows( error_case="When reducing a FeatureCollection with manual windows" ) manual_window = True feat_col_fd_mapping: Dict[str, Tuple[str, FeatureDescriptor]] = {} for (s_names, window), fd_list in self._feature_desc_dict.items(): window = "manual" if manual_window else self._ws_to_str(window) for fd in fd_list: # As a single FeatureDescriptor can have multiple output col names, we # create a unique identifier for each FeatureDescriptor (on which we # will apply set-like operations later on to only retain all the unique # FeatureDescriptors) uuid_str = str(uuid.uuid4()) for output_name in fd.function.output_names: # Reconstruct the feature column name feat_col_name = StridedRolling.construct_output_index( series_keys=s_names, feat_name=output_name, win_str=window ) feat_col_fd_mapping[feat_col_name] = (uuid_str, fd) assert all(fc in feat_col_fd_mapping for fc in feat_cols_to_keep) # Collect (uuid, FeatureDescriptor) for the feat_cols_to_keep fd_subset: List[Tuple[str, FeatureDescriptor]] = [ feat_col_fd_mapping[fc] for fc in feat_cols_to_keep ] # Reduce to unique feature descriptor objects (based on uuid) and create a new # FeatureCollection for their deepcopy's. seen_uuids = set() fds = [] for uuid_str, fd in fd_subset: if uuid_str not in seen_uuids: seen_uuids.add(uuid_str) fds.append(deepcopy(fd)) return FeatureCollection(feature_descriptors=fds)Create a reduced FeatureCollection instance based on
feat_cols_to_keep.For example, this is useful to optimize feature-extraction inference (for your selected features) after performing a feature-selection procedure.
Parameters
feat_cols_to_keep:List[str]- A subset of the feature collection instance its column names.
This corresponds to the columns / names of the output from
calculatemethod that you want to keep.
Returns
FeatureCollection- A new FeatureCollection object, which only withholds the FeatureDescriptors
which constitute the
feat_cols_to_keepoutput.
Note
Some FeatureDescriptor objects may have multiple output-names.
Hence, if you only want to retain a subset of that FeatureDescriptor its feature outputs, you will still get all features as the new FeatureCollection is constructed by applying a filter on de FeatureDescriptor list and we thus not alter these FeatureDescriptor objects themselves.
class FuncWrapper (func, output_names=None, input_type=numpy.ndarray, vectorized=False, **kwargs)-
Expand source code
class FuncWrapper(FrozenClass): """Function wrapper. A function wrapper which takes a numpy array / pandas series as input and returns one or multiple values. It also defines the names of the function outputs, and stores the function its keyword arguments. Parameters ---------- func : Callable The wrapped function. output_names : Union[List[str], str], optional The name of the outputs of the function, by default None. input_type: Union[np.array, pd.Series], optional The input type that the function requires (either np.array or pd.Series), by default np.array. .. Note:: Make sure to only set this argument to pd.Series if the function requires a pd.Series, since pd.Series strided-rolling is significantly less efficient. For a np.array it is possible to create very efficient views, but there is no such thing as a pd.Series view. Thus, for each stroll, a new series is created. vectorized: bool, optional Flag indicating whether `func` should be executed vectorized over all the segmented windows, by default False. .. Info:: A vectorized function should take one or multiple series that each have the shape (nb. segmented windows, window size). For example a vectorized version of `np.max` is ``FuncWrapper(np.max, vectorized=True, axis=1)``. .. Note:: * A function can only be applied in vectorized manner when the required series are REGULARLY sampled (and have the same index in case of multiple required series). * The `input_type` should be `np.ndarray` when `vectorized` is True. It does not make sense to use a `pd.Series`, as the index should be regularly sampled (see requirement above). **kwargs: dict, optional Keyword arguments which will be also passed to the `function` Raises ------ TypeError Raised when the `output_names` cannot be set. """ def __init__( # type: ignore[no-untyped-def] self, func: Callable, output_names: Optional[Union[List[str], str]] = None, input_type: Union[np.ndarray, pd.Series] = np.ndarray, vectorized: bool = False, **kwargs, ): """Create FuncWrapper instance.""" self.func = func self.kwargs: dict = kwargs if isinstance(output_names, list): self.output_names = output_names elif isinstance(output_names, str): self.output_names = [output_names] elif not output_names: self.output_names = [_get_name(func)] else: raise TypeError(f"`output_names` is unexpected type {type(output_names)}") # for backwards compatibility input_type = np.ndarray if input_type is np.array else input_type assert input_type in SUPPORTED_STROLL_TYPES, "Invalid input_type!" assert not ( vectorized & (input_type is not np.ndarray) ), "The input_type must be np.ndarray if vectorized is True!" self.input_type = input_type self.vectorized = vectorized self._freeze() def __repr__(self) -> str: """Return repr string.""" return ( f"{self.__class__.__name__}({_get_name(self.func)}, {self.output_names}," f" {self.kwargs})" ) def __call__(self, *series: Union[np.ndarray, pd.Series]) -> Any: """Call wrapped function with passed data. Parameters --------- *series : Union[np.ndarray, pd.Series] The (multiple) input series for the function. Returns ------- Any The function output for the passed series. """ return self.func(*series, **self.kwargs)Function wrapper.
A function wrapper which takes a numpy array / pandas series as input and returns one or multiple values. It also defines the names of the function outputs, and stores the function its keyword arguments.
Parameters
func:Callable- The wrapped function.
output_names:Union[List[str], str], optional- The name of the outputs of the function, by default None.
input_type:Union[np.array, pd.Series], optional- The input type that the function requires (either np.array or pd.Series), by
default np.array.
Note
Make sure to only set this argument to pd.Series if the function requires a pd.Series, since pd.Series strided-rolling is significantly less efficient. For a np.array it is possible to create very efficient views, but there is no such thing as a pd.Series view. Thus, for each stroll, a new series is created. vectorized:bool, optional-
Flag indicating whether
funcshould be executed vectorized over all the segmented windows, by default False.Info
A vectorized function should take one or multiple series that each have the shape (nb. segmented windows, window size). For example a vectorized version ofnp.maxisFuncWrapper(np.max, vectorized=True, axis=1).Note
- A function can only be applied in vectorized manner when the required series are REGULARLY sampled (and have the same index in case of multiple required series).
- The
input_typeshould benp.ndarraywhenvectorizedis True. It does not make sense to use apd.Series, as the index should be regularly sampled (see requirement above).
**kwargs:dict, optional- Keyword arguments which will be also passed to the
function
Raises
TypeError- Raised when the
output_namescannot be set.
Create FuncWrapper instance.
Ancestors
- tsflex.utils.classes.FrozenClass
Methods
def __call__(self, *series)-
Expand source code
def __call__(self, *series: Union[np.ndarray, pd.Series]) -> Any: """Call wrapped function with passed data. Parameters --------- *series : Union[np.ndarray, pd.Series] The (multiple) input series for the function. Returns ------- Any The function output for the passed series. """ return self.func(*series, **self.kwargs)Call wrapped function with passed data.
Parameters
*series:Union[np.ndarray, pd.Series]- The (multiple) input series for the function.
Returns
Any- The function output for the passed series.
class StridedRollingFactory-
Expand source code
class StridedRollingFactory: """Factory class for creating the appropriate StridedRolling segmenter.""" _datatype_to_stroll = { DataType.TIME: TimeStridedRolling, DataType.SEQUENCE: SequenceStridedRolling, } @staticmethod def get_segmenter( # type: ignore[no-untyped-def] data: Union[pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]]], window: Union[int, float, pd.Timedelta], strides: Optional[List[Union[int, float, pd.Timedelta]]], **kwargs, ) -> StridedRolling: """Get the appropriate StridedRolling instance for the passed data. The returned instance will be determined by the data its index type Parameters ---------- data : Union[pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]]] The data to segment. window : Union[int, float, pd.Timedelta] The window size to use for the segmentation. strides : Union[List[Union[int, float, pd.Timedelta]], None] The stride(s) to use for the segmentation. **kwargs : dict, optional Additional keyword arguments, see the `StridedRolling` its documentation for more info. .. Note:: When passing `time-based` data, but **int**-based window & stride params, the strided rolling will be `TimeIndexSampleStridedRolling`. This class **assumes** that **all data series** _roughly_ have the **same sample frequency**, as the windows and strides are interpreted in terms of **number of samples**. Raises ------ ValueError When incompatible data & window-stride data types are passed (e.g. time window-stride args on sequence data-index). Returns ------- StridedRolling The constructed StridedRolling instance. """ data_dtype = AttributeParser.determine_type(data) if strides is None: args_dtype = AttributeParser.determine_type(window) else: args_dtype = AttributeParser.determine_type([window] + strides) if window is None or data_dtype.value == args_dtype.value: return StridedRollingFactory._datatype_to_stroll[data_dtype]( data, window, strides, **kwargs ) elif data_dtype == DataType.TIME and args_dtype == DataType.SEQUENCE: # Note: this is very niche and thus requires advanced knowledge assert isinstance(window, int) if strides is not None: assert isinstance(strides, list) and all( isinstance(s, int) for s in strides ) return TimeIndexSampleStridedRolling(data, window, strides, **kwargs) elif data_dtype == DataType.SEQUENCE and args_dtype == DataType.TIME: raise ValueError("Cannot segment a sequence-series with a time window") # This should never happen raise ValueError( f"Cannot segment data of type {data_dtype} with window-stride of type {args_dtype}" )Factory class for creating the appropriate StridedRolling segmenter.
Static methods
def get_segmenter(data, window, strides, **kwargs)-
Expand source code
@staticmethod def get_segmenter( # type: ignore[no-untyped-def] data: Union[pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]]], window: Union[int, float, pd.Timedelta], strides: Optional[List[Union[int, float, pd.Timedelta]]], **kwargs, ) -> StridedRolling: """Get the appropriate StridedRolling instance for the passed data. The returned instance will be determined by the data its index type Parameters ---------- data : Union[pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]]] The data to segment. window : Union[int, float, pd.Timedelta] The window size to use for the segmentation. strides : Union[List[Union[int, float, pd.Timedelta]], None] The stride(s) to use for the segmentation. **kwargs : dict, optional Additional keyword arguments, see the `StridedRolling` its documentation for more info. .. Note:: When passing `time-based` data, but **int**-based window & stride params, the strided rolling will be `TimeIndexSampleStridedRolling`. This class **assumes** that **all data series** _roughly_ have the **same sample frequency**, as the windows and strides are interpreted in terms of **number of samples**. Raises ------ ValueError When incompatible data & window-stride data types are passed (e.g. time window-stride args on sequence data-index). Returns ------- StridedRolling The constructed StridedRolling instance. """ data_dtype = AttributeParser.determine_type(data) if strides is None: args_dtype = AttributeParser.determine_type(window) else: args_dtype = AttributeParser.determine_type([window] + strides) if window is None or data_dtype.value == args_dtype.value: return StridedRollingFactory._datatype_to_stroll[data_dtype]( data, window, strides, **kwargs ) elif data_dtype == DataType.TIME and args_dtype == DataType.SEQUENCE: # Note: this is very niche and thus requires advanced knowledge assert isinstance(window, int) if strides is not None: assert isinstance(strides, list) and all( isinstance(s, int) for s in strides ) return TimeIndexSampleStridedRolling(data, window, strides, **kwargs) elif data_dtype == DataType.SEQUENCE and args_dtype == DataType.TIME: raise ValueError("Cannot segment a sequence-series with a time window") # This should never happen raise ValueError( f"Cannot segment data of type {data_dtype} with window-stride of type {args_dtype}" )Get the appropriate StridedRolling instance for the passed data.
The returned instance will be determined by the data its index type
Parameters
data:Union[pd.Series, pd.DataFrame, List[Union[pd.Series, pd.DataFrame]]]- The data to segment.
window:Union[int, float, pd.Timedelta]- The window size to use for the segmentation.
strides:Union[List[Union[int, float, pd.Timedelta]], None]- The stride(s) to use for the segmentation.
**kwargs:dict, optional- Additional keyword arguments, see the
StridedRollingits documentation for more info.
Note
When passing
time-baseddata, but int-based window & stride params, the strided rolling will beTimeIndexSampleStridedRolling. This class assumes that all data series roughly have the same sample frequency, as the windows and strides are interpreted in terms of number of samples.Raises
ValueError- When incompatible data & window-stride data types are passed (e.g. time window-stride args on sequence data-index).
Returns
StridedRolling- The constructed StridedRolling instance.